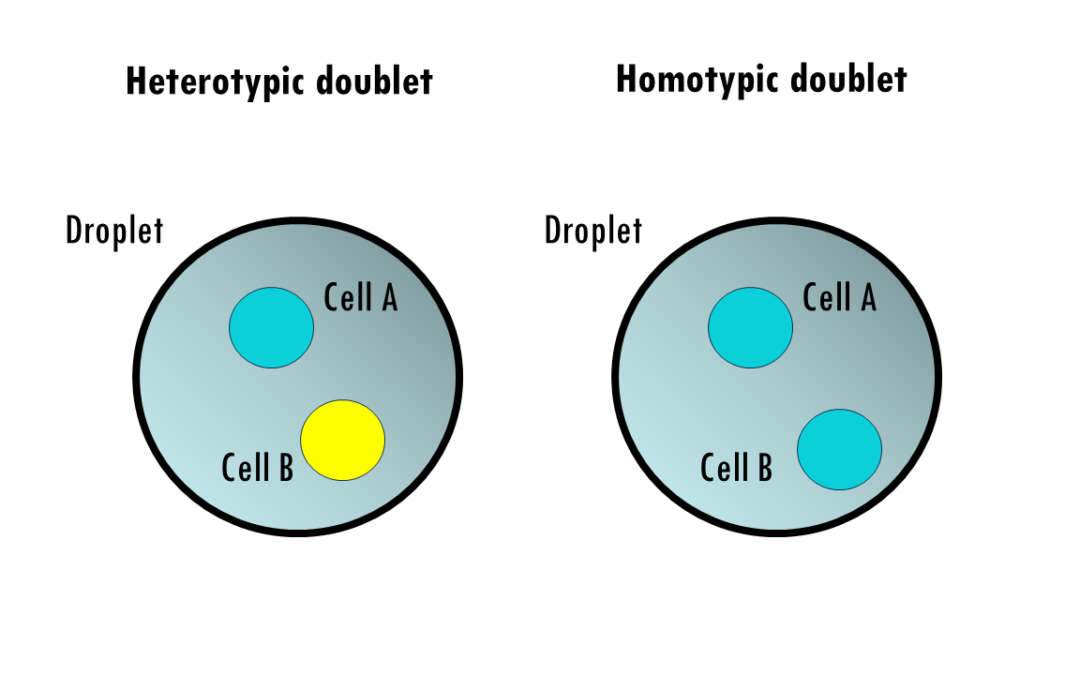
by Biostatsquid | Aug 18, 2024 | scRNAseq, Tutorials
A step-by-step easy R tutorial to detect and remove doublets using scDblFinder in R In this easy, step-by-step scDblFinder tutorial you will learn how to detect and remove doublets from scRNAseq data in R, using the R package (you guessed it!) scDblFinder . First...

by Biostatsquid | Mar 11, 2024 | scRNAseq, Tutorials
A step-by-step easy R tutorial for cell type annotation with SingleR In this easy, step-by-step tutorial you will learn how to to do cell type annotation with the R package SingleR. SingleR is a popular reference-based automatic cell type annotation tool used to...
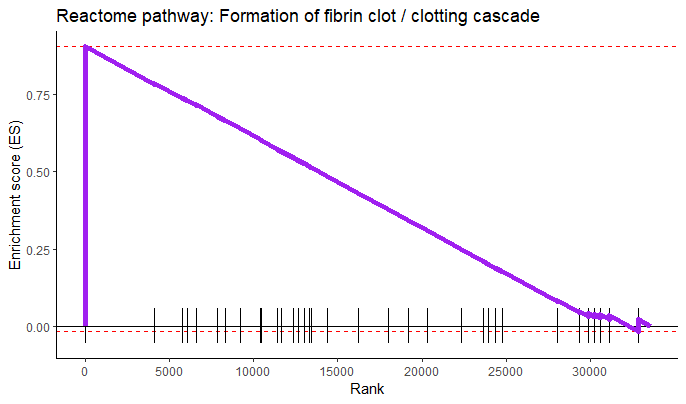
by Biostatsquid | Sep 4, 2023 | scRNAseq, Statistics in R, Tutorials
A step-by-step R tutorial on performing Gene Set Enrichment Analysis with R package fgsea In this tutorial you will learn how to conduct Gene Set Enrichment Analysis (GSEA) using R-package fgsea. This package implements an algorithm for fast gene set enrichment...
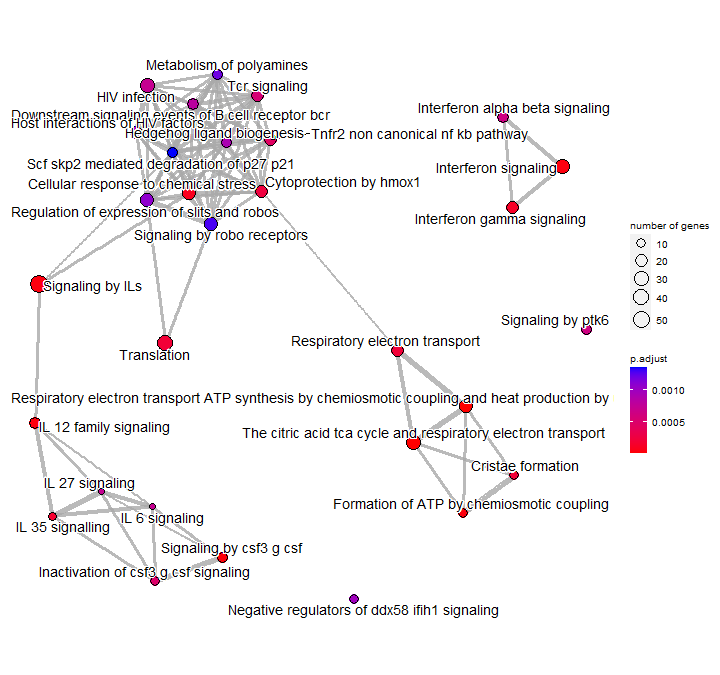
by Biostatsquid | Jul 9, 2023 | scRNAseq, Tutorials
A step-by-step R tutorial on performing pathway enrichment analysis with clusterProfiler in R Note! This is an updated version of ORA analysis with clusterProfiler which doesn’t correspond exactly to the video tutorial! Click here for the old version. Table of...
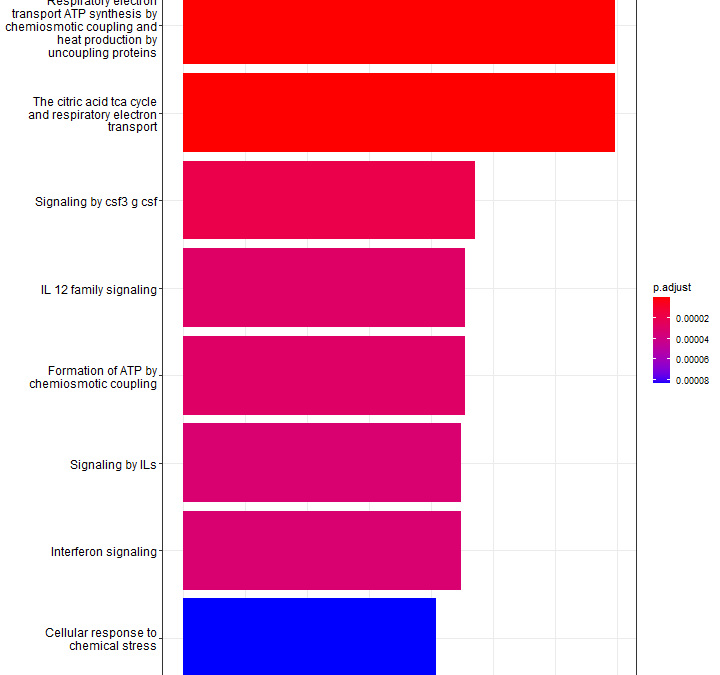
by Biostatsquid | Jul 9, 2023 | scRNAseq, Tutorials
A step-by-step R tutorial on visualising pathway enrichment analysis results with clusterProfiler Table of contents 5 Before you start 5 Step-by-step tutorial 9 Creating an enrichResult object 9 Barplots 9 Dotplots 9 Cnetplot 9 Heatplot 9 Treeplot 9 Enrichment map 9...
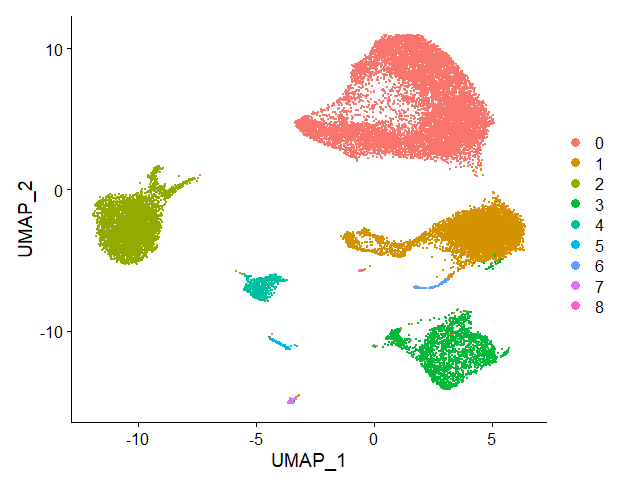
by Biostatsquid | Jan 10, 2023 | scRNAseq, Tutorials
R TUTORIAL: Pre-process your scRNAseq data with this easy, step-by-step pipeline Table of contents 5 Before you start 5 scRNAseq pre-processing pipeline: complete script 5 Step-by-step guide to analyse your data with Seurat 9 Step 1: Set up your script 9 Step 2:...








