by Biostatsquid | Dec 14, 2025 | Learning, scRNAseq
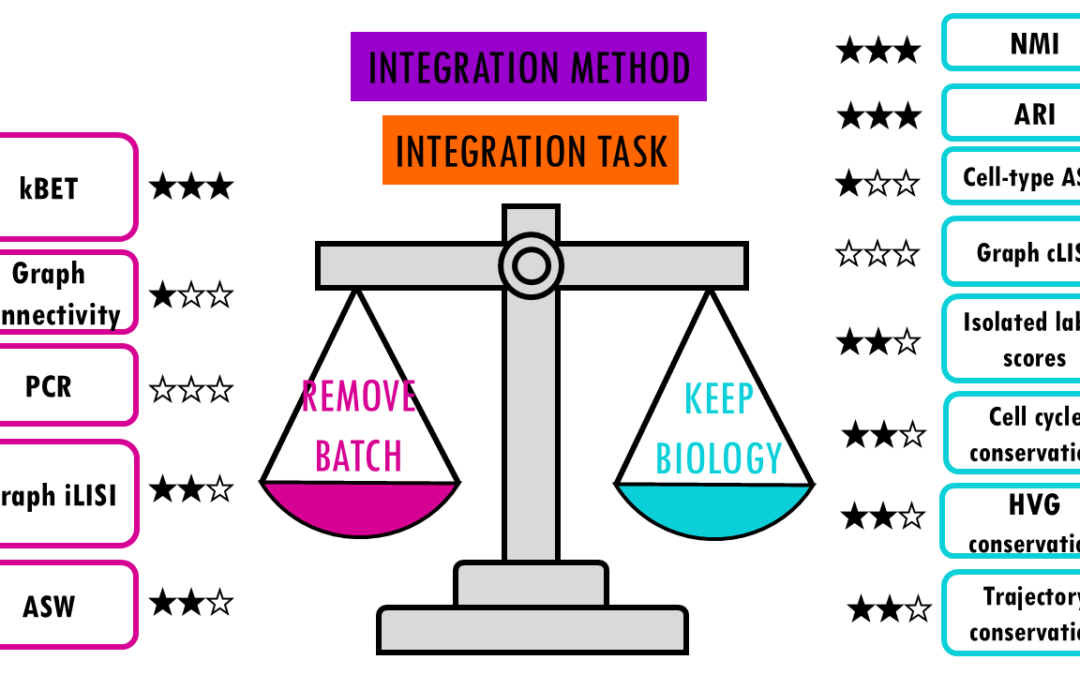
Comparing top integration methods for scRNAseq data When we want to combine multiple scRNA-seq datasets to answer bigger questions, we encounter batch effects – unwanted technical variations that arise from differences in how the experiments were performed....
by Biostatsquid | Nov 30, 2025 | Learning, scRNAseq
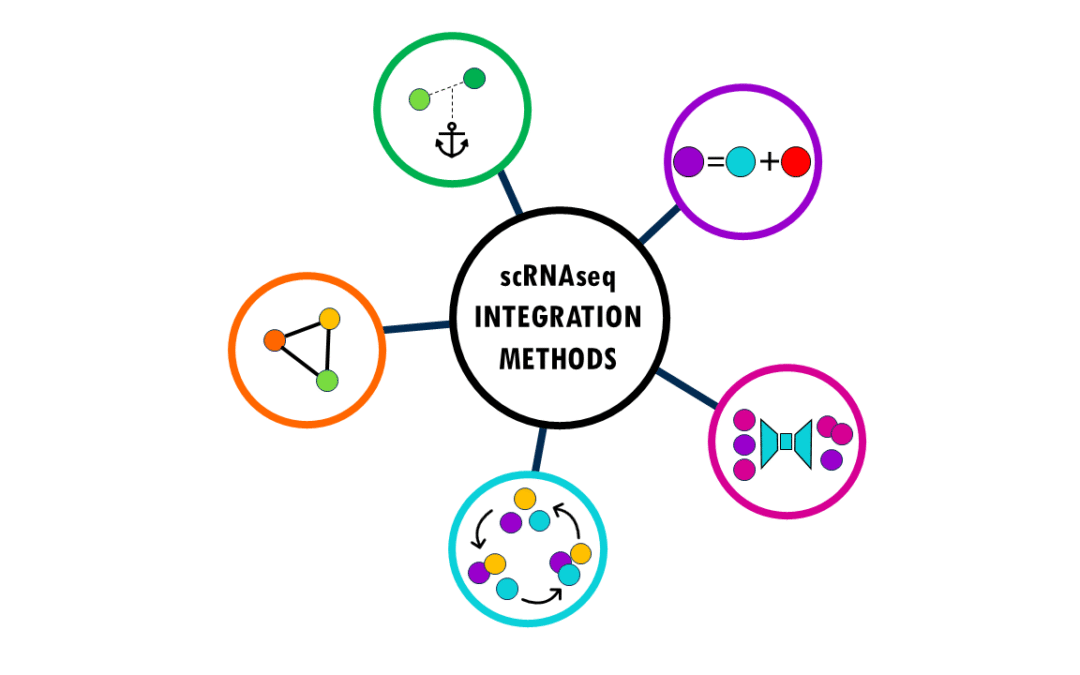
An overview of the most popular integration methods for single-cell data Single-cell RNA sequencing (scRNA-seq) has revolutionized our understanding of biology by allowing us to measure gene expression in individual cells rather than bulk tissue samples. This...
by Biostatsquid | Nov 15, 2025 | Learning, RNAseq, scRNAseq, scRNAseq, Tutorials
MSigDB gene sets: easy msigdbr in R MSigDB gene sets: easy msigdbr in R Welcome to this comprehensive guide on MSigDB (Molecular Signatures Database) and the msigdbr R package! If you’ve ever wondered which gene sets to use for your pathway enrichment analysis, or...
by Biostatsquid | Jul 15, 2025 | RNAseq, scRNAseq, Tutorials, Uncategorized
Hey! You’re looking at an old post. Newer version here: clusterProfiler tutorial in R R TUTORIAL: Perform pathway enrichment analysis with clusterProfiler() in R Table of contents 5 Before you start 5 Step-by-step clusterProfiler tutorial 9 Step 0: Set up your...

by Biostatsquid | Apr 27, 2025 | Learning, RNAseq, scRNAseq
Understanding the structure of Seurat objects version 5 – step-by-step simple explanation! If you’ve worked with single-cell RNAseq data, you’ve probably heard about Seurat. In this blogpost, we’ll cover the the Seurat object structure,in...
by Biostatsquid | Apr 24, 2025 | scRNAseq, Statistics
SCTransform (Single-Cell Transform) is a normalization method primarily used in scRNA-seq data analysis. It was developed to address limitations in standard normalization approaches when dealing with single-cell data. You can check how to apply SCTransform on your...





