by Biostatsquid | Nov 15, 2025 | Learning, RNAseq, scRNAseq, scRNAseq, Tutorials
MSigDB gene sets: easy msigdbr in R MSigDB gene sets: easy msigdbr in R Welcome to this comprehensive guide on MSigDB (Molecular Signatures Database) and the msigdbr R package! If you’ve ever wondered which gene sets to use for your pathway enrichment analysis, or...
by Biostatsquid | Sep 21, 2025 | scRNAseq, Tutorials, Visualisation
ComplexHeatmap Tutorial: create publication-ready heatmaps in R ComplexHeatmap Tutorial: create publication-ready heatmaps in R In this easy step-by-step tutorial we will learn how to create and customise a heatmap with R package ComplexHeatmap. ComplexHeatmap is one...
by Biostatsquid | Jul 15, 2025 | scRNAseq, Tutorials
Hey! You’re looking at an old post. Newer version here: fgsea tutorial in R A step-by-step R tutorial on performing Gene Set Enrichment Analysis with R package fgsea In this tutorial you will learn how to conduct Gene Set Enrichment Analysis (GSEA) using...

by Biostatsquid | May 5, 2025 | scRNAseq, Tutorials
A step-by-step easy R tutorial to preprocess scRNAseq data with Seurat v5 In this easy, step-by-step tutorial you will learn how a Seurat object is structured and how to preprocess scRNAseq data using the standard workflow with Seurat v5. This is a hands-on...
by Biostatsquid | Aug 25, 2024 | scRNAseq, Tutorials
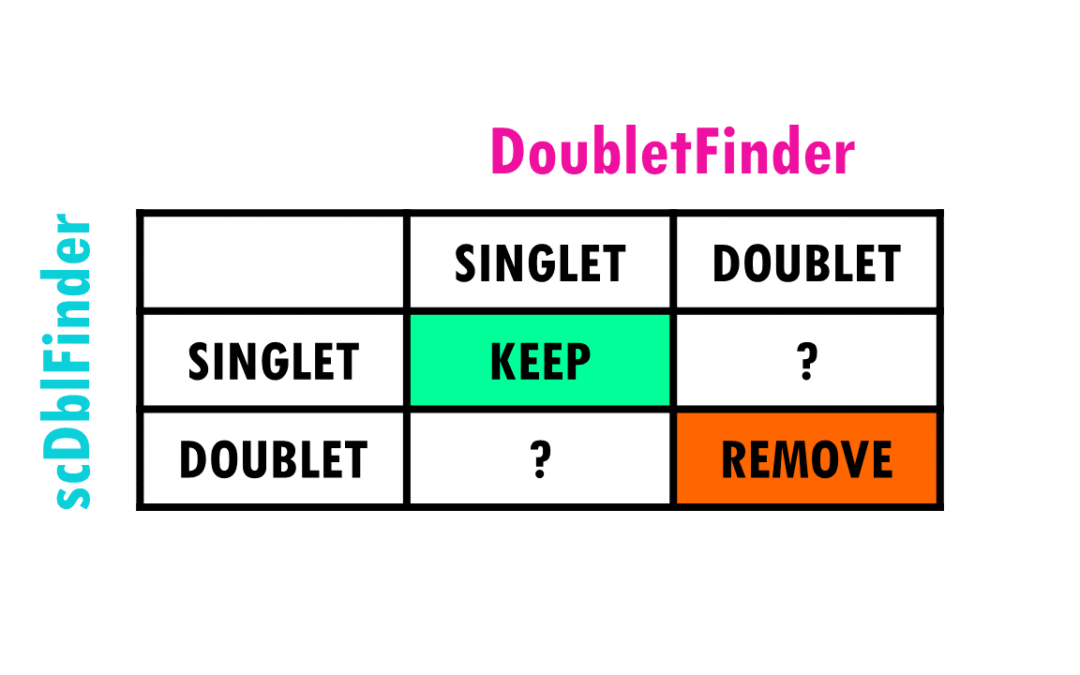
A step-by-step easy R tutorial to detect and remove doublets from scRNAseq data In this easy, step-by-step tutorial you will learn how to do some general doublet QC and remove doublets from our Seurat object in R for scRNAseq data. This is part 3 of my tutorial series...
by Biostatsquid | Aug 18, 2024 | scRNAseq, Tutorials
A step-by-step easy R tutorial to detect and remove doublets using DoubletFinder in R In this easy, step-by-step tutorial you will learn how to detect and remove doublets from scRNAseq data in R, using the R package DoubletFinder. First things first. What are...




