Hey! You’re looking at an old post. Newer version here: fgsea tutorial in R
A step-by-step R tutorial on performing Gene Set Enrichment Analysis with R package fgsea
In this tutorial you will learn how to conduct Gene Set Enrichment Analysis (GSEA) using R-package fgsea. This package implements an algorithm for fast gene set enrichment analysis.
Following this easy, step-by-step tutorial, you will find out how to:
-
- Install and start fgsea()
- Prepare your dataset to perform GSEA
- Set the analysis parameters and run the analysis.
- View the GSEA results and get some nice plots!
If you are more of a video-based learner, you can check out my Youtube video on how to perform GSEA with fgsea() in R. Otherwise, just keep reading! And remember you can just copy the code by clicking on the top right corner of the code snippets in this post.
Let’s dive in!
Check out my Youtube video to follow this fgsea R tutorial with me!
Before you start…
In this tutorial we will learn how to perform Gene Set Enrichment Analysis on differential gene expression results. We will use the R package fgsea() for fast preranked gene set enrichment analysis (GSEA).
If you are not that familiar with GSEA, you might want to check my other post on GSEA – simply explained! I go over the basic concepts behind GSEA algorithms and how to interpret the results.
If you are a beginner in R, don’t be overwhelmed! This tutorial will go step-by-step and I will explain (almost!) every line of code so you know what is happening at each point of the workflow.
For this tutorial you will need R, or Rstudio, and you will need to install fgsea.
Install fgsea()
You can read more about fgsea() package here. To install fgsea, use the following command:
BiocManager::install("fgsea")Once installed, we can load the package:
library("fgsea")Set up your environment
It’s good practice to clear up your environment before you load in any data. This is the general structure I like to keep in all my scripts:
# ----------------------
# GSEA tutorial
# ----------------------
# Setting up environment ===================================================
# Clean environment
rm(list = ls(all.names = TRUE)) # will clear all objects including hidden objects
gc() # free up memory and report the memory usage
options(max.print = .Machine$integer.max, scipen = 999, stringsAsFactors = F, dplyr.summarise.inform = F) # avoid truncated output in R console and scientific notation
# Set seed
set.seed(123456)
# Set project library
.libPaths('C:/Users/laura/Documents/Biostatsquid/Scripts/R4.2.3')
# Loading relevant libraries
library(tidyverse) # includes ggplot2, for data visualisation. dplyr, for data manipulation.
library(RColorBrewer) # for a colourful plot
library(fgsea)
# Set relevant paths
list.files()
in_path <- "Datasets/"
out_path <- "PEA/Results/"What do I need to run fgsea?
Essentially, there are two things you need to perform preranked gene set enrichment analysis with fgsea.
The fgsea package takes in two main arguments.
The first one is pathways: a list of gene sets or pathways to check
The second one is stats: a named vector of our genes of interest we want to perform GSEA on. The gene names must be the same as the ones in pathways! So make sure you are using the same nomenclature (i.e., gene IDs or Ensemble IDs).
That’s it!
There are additional parameters you may be interested in such as:
- minSize: minimal size of a gene set to test (all pathways below the threshold are excluded)
- maxSize: the same, but a maximum threshold.
- scoreType: the default is ‘std’, where the enrichment score is computed normally, but you can also use one-tailed tests if you are only interested in positive (‘pos’) enrichment – so only pathways that are overrepresented – or negative (‘neg’) enrichment – to only get pathways that are underrepresented in your list of genes.
1. Prepare your gene sets / pathways
As we already saw in the Pathway Enrichment Analysis with clusterProfiler() tutorial, we need to prepare a list of pathways.
1. Get your gene set .gmt file. For example, you can download it from:
2. Filter the .gmt file and convert it to a format accepted by fgsea. As I explain in my previous post, we need to use a set of background genes that contain only the genes present in our dataframe, to avoid bias. Note that this is done automatically by the fgsea package, so no need to do it yourself (which is why I commented out the line that does the filtering). You can use the following code:
# Function: Adjacency matrix to list -------------------------
matrix_to_list <- function(pws){
pws.l <- list()
for (pw in colnames(pws)) {
pws.l[[pw]] <- rownames(pws)[as.logical(pws[, pw])]
}
return(pws.l)
}
# Get all the genes in your dataset and assign them to my_genes
my_genes <- df$gene_symbol
# Download gene sets .gmt files
#https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp
# Copy the .gmt file to your folder, in my case, its 'PEA/Background_genes/'
# Then read in the .gmt file
gmt_file <- gmtPathways("PEA/Background_genes/c2.cp.kegg.v7.5.1.symbols.gmt")
hidden <- unique(unlist(gmt))
# Convert gmt file to a matrix with the genes as rows and for each go annotation (columns) the values are 0 or 1
mat <- matrix(NA, dimnames = list(hidden, names(gmt)),
nrow = length(hidden), ncol = length(gmt))
for (i in 1:dim(mat)[2]){
mat[,i] <- as.numeric(hidden %in% gmt[[i]])
}
#Subset to the genes that are present in our data to avoid bias
#hidden1 <- intersect(genes_in_data, hidden)
#mat <- mat[hidden1, colnames(mat)[which(colSums(mat[hidden1,])>5)]] # filter for gene sets with more than 5 genes annotated
# And get the list again using the function we previously defined
final_list <- matrix_to_list(mat)
Squidtip
In my old version (and in the video), I used to download the .gmt files with the pathways manually from the msigdb database. Now, it’s so much easier! You can use the R package msigdbr to download whichever pathways you’re interested in. If you are unsure which one is right for you, check out this other post where I explain the different pathways and gene sets available in msigdb.
Alternatively, if you want to run GSEA on many gene sets (e.g., KEGG, Reactome, Gene Ontology…), you can use a function:
# Set relevant paths
list.files()
in_path <- "Datasets/"
out_path <- "PEA/Results/"
bg_path <- "PEA/Background_genes/"
# Functions ===================================================
## Function: Adjacency matrix to list -------------------------
matrix_to_list <- function(pws){
pws.l <- list()
for (pw in colnames(pws)) {
pws.l[[pw]] <- rownames(pws)[as.logical(pws[, pw])]
}
return(pws.l)
}
## Function: prepare_gmt --------------------------------------
prepare_gmt <- function(gmt_file, genes_in_data, savefile = FALSE){
# for debug
#file <- gmt_files[1]
#genes_in_data <- df$gene_symbol
# Read in gmt file
gmt <- gmtPathways(gmt_file)
hidden <- unique(unlist(gmt))
# Convert gmt file to a matrix with the genes as rows and for each go annotation (columns) the values are 0 or 1
mat <- matrix(NA, dimnames = list(hidden, names(gmt)),
nrow = length(hidden), ncol = length(gmt))
for (i in 1:dim(mat)[2]){
mat[,i] <- as.numeric(hidden %in% gmt[[i]])
}
#Subset to the genes that are present in our data to avoid bias
hidden1 <- intersect(genes_in_data, hidden)
mat <- mat[hidden1, colnames(mat)[which(colSums(mat[hidden1,])>5)]] # filter for gene sets with more than 5 genes annotated
# And get the list again
final_list <- matrix_to_list(mat) # for this we use the function we previously defined
if(savefile){
saveRDS(final_list, file = paste0(gsub('.gmt', '', gmt_file), '_subset_', format(Sys.time(), '%d%m'), '.RData'))
}
print('Wohoo! .gmt conversion successfull!:)')
return(final_list)
}
# Analysis ====================================================
## 1. Read in data -----------------------------------------------------------
list.files(in_path)
df <- read.csv(paste0(in_path, 'severevshealthy_degresults.csv'), row.names = 1)
## 2. Prepare background genes -----------------------------------------------
# Download gene sets .gmt files
#https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp
# For GSEA
# Filter out the gmt files for KEGG, Reactome and GOBP
my_genes <- df$gene_symbol
list.files(bg_path)
gmt_files <- list.files(path = bg_path, pattern = '.gmt', full.names = TRUE)
gmt_files
bg_genes <- prepare_gmt(gmt_files[2], genes_in_data, savefile = FALSE)This way, if I want to use a different background gene set, I can just change the parameters of my custom function, prepare_gmt():
# gmt_files is just a list of filenames of all my .gmt files (I have 3):
> gmt_files
[1] "PEA/Background_genes/c2.cp.kegg.v7.5.1.symbols.gmt" "PEA/Background_genes/c2.cp.reactome.v7.5.1.symbols.gmt"
[3] "PEA/Background_genes/c5.go.bp.v7.5.1.symbols.gmt# You can use a list of filenames, and just change [1], [2], [3]...
bg_genes <- prepare_gmt(gmt_files[1], my_genes, savefile = FALSE)
# Or write it out
bg_genes <- prepare_gmt("PEA/Background_genes/c5.go.bp.v7.5.1.symbols.gmt", my_genes, savefile = FALSE)Squidtip
Did you know? The fgsea package already provides a function that returns a list of Reactome pathways for a given vector of Entrez gene IDs.
Just call reactomePathways(genes).
Your bg_genes object should be a list with your pathway or gene set names, and the genes that make up each pathway.
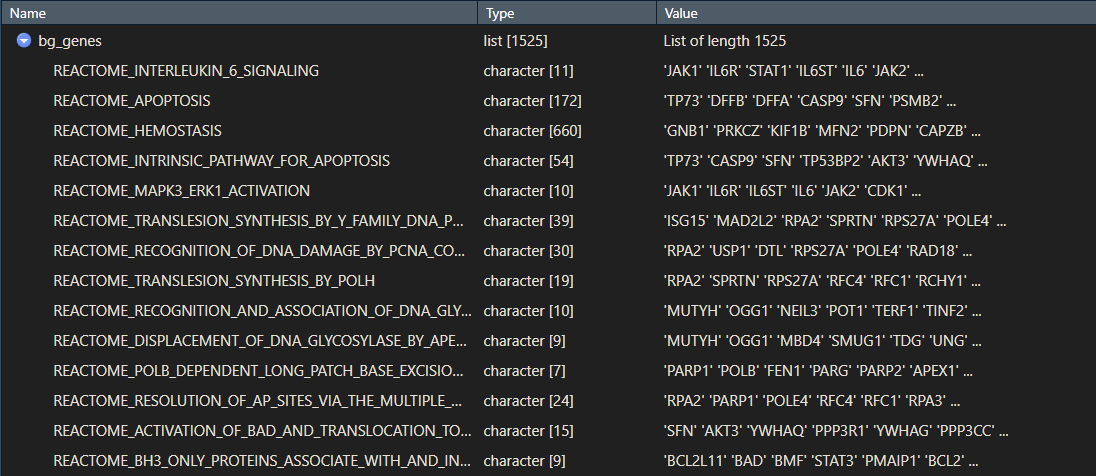
2. Prepare your ranked list of genes
We would like to perform GSEA on our differentially expressed genes. Unlike pathway enrichment analysis, GSEA provides the advantage of not needing to subset our gene list. We can provide all the genes that resulted from our differential expression analysis. Even genes that were not significantly differentially expressed.
We just need to rank our genes first.
The way it works is, you need to give fgsea a named vector with the rankings and the genes symbols as names.
If you are not very clear on this, you might want to check out my post on GSEA – simply explained!
Squidtip
How do you rank your genes?
There are many ways of ranking your genes, so you need to decide based on your analysis. Here, I use the sign(df$log2fc)*(-log10(df$pval))) as a ranking metric.
This way, the magnitude of the ranking will be given by the p value (since I use -log10(), the more significant a gene is differentially expressed, the larger that value will be). The sign of the ranking (positive or negative) will be given by the fold-change: upregulated genes will have positive rankings, downregulated genes will have negative rankings.
rankings <- sign(df$log2fc)*(-log10(df$pval)) # we will use the signed p values from spatial DGE as ranking
names(rankings) <- df$gene_symbol # genes as names#Easy! We’ve just created our ranked list of genes.
You can have a look at the values:
> head(rankings)
MIR1302-2HG FAM138A OR4F5 AL627309.1 AL627309.3 AL627309.2
0.000061381930 -0.000005164293 -0.000005164293 -0.154354567687 -0.000005164293 -0.000005164293
> rankings <- sort(rankings, decreasing = TRUE) # sort genes by ranking
> plot(rankings)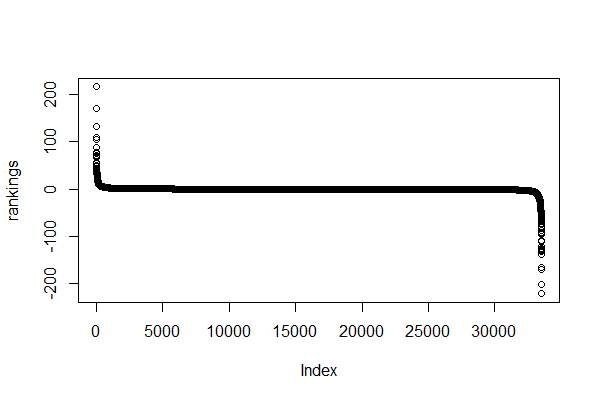
Yes, it’s an ugly plot, I know.
In the x axis, we see our genes (each dot represents a gene) – they are indexed so instead of gene symbols we just see numbers from 1 to 33538.
In the y axis, we see the ranking (sign(df$log2fc)*(-log10(df$pval)))) for each gene.
Squidtip
Another note on rankings… You can just use the log2(fold-change) as a ranking – but it will not take into account genes with a large fold change but non-significant. However, if you have already selected your significant genes, this may be a good option for you.
Most genes have a ranking value between -100 and 100.
A few ones seem to have higher rankings. Let’s have a closer look…
> max(rankings)
[1] Inf
> min(rankings)
[1] -InfIt seems like some genes have a ranking of infinity. This is due to the fact that some p-values were really small, so computing -log10(p-value) returned infinity.
fgsea() does not accept non-finite numbers as rankings, so we need to fix that.
In this case, we will just convert them to really large (or small) values:
# Some genes have such low p values that the signed pval is +- inf, we need to change it to the maximum * constant to avoid problems with fgsea
max_ranking <- max(rankings[is.finite(rankings)])
min_ranking <- min(rankings[is.finite(rankings)])
rankings <- replace(rankings, rankings > max_ranking, max_ranking * 10)
rankings <- replace(rankings, rankings < min_ranking, min_ranking * 10)
rankings <- sort(rankings, decreasing = TRUE) # sort genes by rankingBy replacing the infinite values by ten times the maximum or minimum ranking, we avoid errors when running fgsea().
Now, if we plot the rankings:
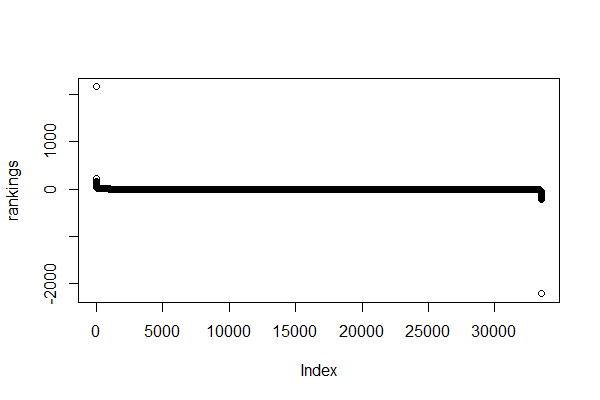
> max(rankings)
[1] 2152.518
> min(rankings)
[1] -2206.971You can also check the rankings of a specific set of genes and a nicer plot with ggplot.
Let’s have a look at the first 50 genes (if you don’t have many genes you can try with all but since I have 33538 I don’t want it to crash – again).
ggplot(data.frame(gene_symbol = names(rankings)[1:50], ranks = rankings[1:50]), aes(gene_symbol, ranks)) +
geom_point() +
theme_classic() +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))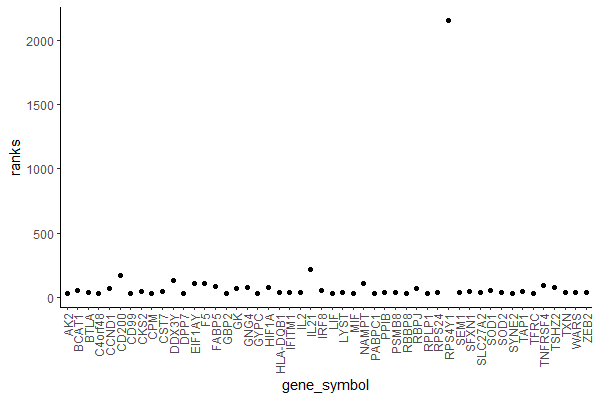
3. Run GSEA
## 4. Run GSEA ---------------------------------------------------------------
# Easy peasy! Run fgsea with the pathways
GSEAres <- fgsea(pathways = bg_genes, # List of gene sets to check
stats = rankings,
scoreType = 'std', # in this case we have both pos and neg rankings. if only pos or neg, set to 'pos', 'neg'
minSize = 10,
maxSize = 500,
nproc = 1) # for parallelisationYou might get a warning saying:
Warning message:
In preparePathwaysAndStats(pathways, stats, minSize, maxSize, gseaParam, :
There are ties in the preranked stats (48.8% of the list).
The order of those tied genes will be arbitrary, which may produce unexpected results.As it states there, there are many genes with ‘tied’ scores or rankings. Genes with the same score will just be ordered at random by the algorithm, and might slightly affect your results when computing the Enrichment Score. This just means that if genes #15 # 16 and #17 have a ranking of ’10’, they will be ordered at random (#17 #15 #16).
It shouldn’t have a very big effect on your results. In my case, it mostly affects genes with a ranking near 0 (they were not significant genes).
Squidtip
About the scoreType… If you have only positive rankings, you can change this to ‘pos’. For example, if you are using the absolute value of logFC. It is not very common, but you can read more about use cases here.
> head(GSEAres)
pathway pval padj log2err
1: REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM 0.3633218 0.9085975 0.08862611
2: REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT 0.4397394 0.9085975 0.11573445
3: REACTOME_ABC_TRANSPORTERS_IN_LIPID_HOMEOSTASIS 0.9746835 0.9856000 0.05773085
4: REACTOME_ABC_TRANSPORTER_DISORDERS 0.3471338 0.9085975 0.13077714
5: REACTOME_ABERRANT_REGULATION_OF_MITOTIC_EXIT_IN_CANCER_DUE_TO_RB1_DEFECTS 0.9820051 0.9882600 0.05809836
6: REACTOME_ABERRANT_REGULATION_OF_MITOTIC_G1_S_TRANSITION_IN_CANCER_DUE_TO_RB1_DEFECTS 0.5102564 0.9085975 0.09167952
ES NES size leadingEdge
1: -0.7980657 -1.1339616 10 ABCB1
2: 0.4951437 0.9782602 103 PSMB8,SEM1,PSMB9,PSMB3,PSMA6,PSME1,...
3: 0.3593687 0.5781693 18 PEX3,ABCG1,APOA1,ABCG8,ABCA3,ABCA10
4: 0.5572234 1.0724269 78 PSMB8,SEM1,PSMB9,PSMB3,PSMA6,PSME1,...
5: 0.3580019 0.5778174 20 ANAPC5,ANAPC2,CDC26,CDC27,UBE2C,RB1,...
6: 0.6152041 0.9756658 17 CCND1,CCND3,CCNE14. Visualise your GSEA results
Let’s have a look at our top 6 enriched pathways, ordered by p-value or p-adjusted value.
## 6. Check results ------------------------------------------------------
# Top 6 enriched pathways (ordered by p-val)
head(GSEAres[order(pval), ])How many significant pathways did we get?
> sum(GSEAres[, padj < 0.01])
[1] 0
> sum(GSEAres[, pval < 0.01])
[1] 8Clearly, our results are not great. There are only 8 significant pathways (p-value < 0.05), but when correcting for multiple testing, we have no significant pathways (p-adjusted values < 0.05).
Oh, well. For the sake of this tutorial, let’s keep going! Hopefully your results will be prettier than mine…
Let’s plot our top pathways. I ordered them by p-value because my pathway enrichment analysis did not yield very interesting results… but you can change it to p-adjusted values if you would like to be more restrictive and correct for multiple testing…
topPathwaysUp <- GSEAres[ES > 0][head(order(pval), n = number_of_top_pathways_up), pathway]
topPathwaysDown <- GSEAres[ES < 0][head(order(pval), n = number_of_top_pathways_down), pathway]
topPathways <- c(topPathwaysUp, rev(topPathwaysDown))
#pdf(file = paste0(filename, '_gsea_top30pathways.pdf'), width = 20, height = 15)
plotGseaTable(bg_genes[topPathways], stats = rankings, fgseaRes = GSEAres, gseaParam = 0.5)
#dev.off()
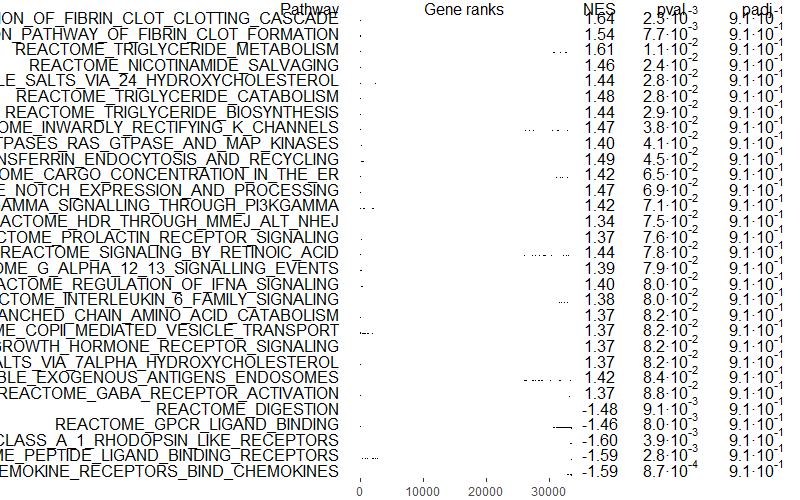
You can also select only independent pathways, removing redundancies or similar pathways with the function collapsePathways():
# Select only independent pathways, removing redundancies/similar pathways
collapsedPathways <- collapsePathways(GSEAres[order(pval)][pval < 0.05], bg_genes, rankings)
mainPathways <- GSEAres[pathway %in% collapsedPathways$mainPathways][order(-NES), pathway]
#pdf(file = paste0('GSEA/Selected_pathways/', paste0(filename, background_genes, '_gsea_mainpathways.pdf')), width = 20, height = 15)
plotGseaTable(bg_genes[mainPathways], rankings, GSEAres, gseaParam = 0.5)
#dev.off()
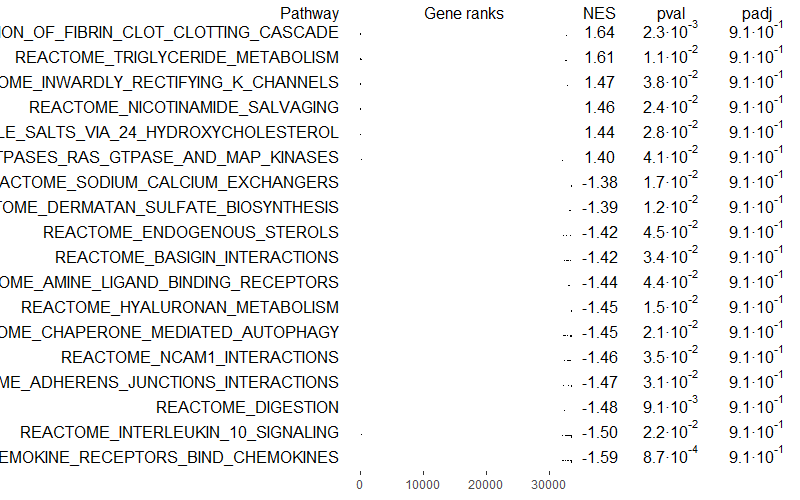
Squidtip
If you’d like to export the tables, just uncomment the 2 lines above. You can also export to .png, or other formats:
png(file = paste0(filename, ‘_gsea_mainpathways.png’), width = 1500, height = 800)
plotGseaTable(bg_genes[mainPathways], rankings, GSEAres, gseaParam = 0.5)
dev.off()
Now let’s plot the ES of the most significantly enriched pathway.
# plot the most significantly enriched pathway
plotEnrichment(bg_genes[[head(GSEAres[order(padj), ], 1)$pathway]],
rankings) +
labs(title = head(GSEAres[order(padj), ], 1)$pathway)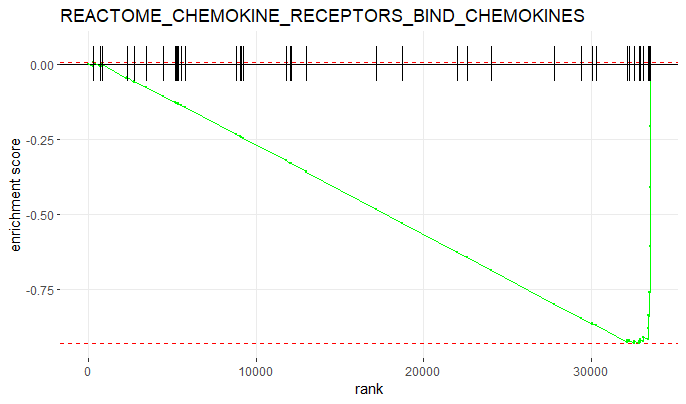
You can also edit the plot and make it prettier;)
plotEnrichment(bg_genes[['REACTOME_FORMATION_OF_FIBRIN_CLOT_CLOTTING_CASCADE']],
rankings) +
labs(title = 'Reactome pathway: Formation of fibrin clot / clotting cascade') +
theme_classic() +
scale_x_continuous('Rank', breaks = seq(0, 32000, 5000)) +
scale_y_continuous('Enrichment score (ES)') +
geom_line(col = 'purple', linewidth = 2)
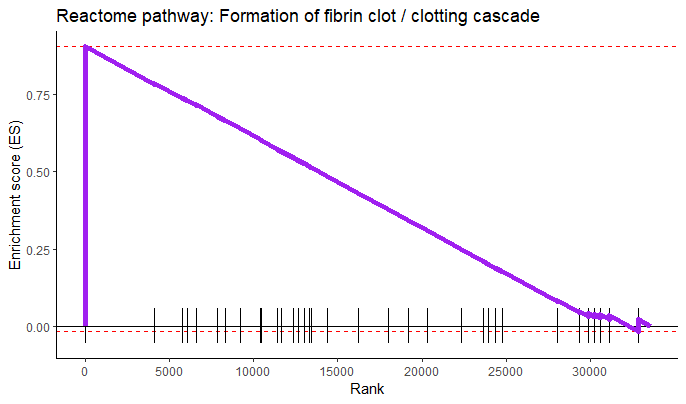
5. Save your results
## 5. Save the results -----------------------------------------------
name_of_comparison <- 'severevshealthy'
background_genes <- 'gobp'
filename <- paste0(out_path, 'GSEA/', name_of_comparison, '_', background_genes)
#saveRDS(GSEAres, file = paste0(filename, '_gsea_results.RDS'))
#data.table::fwrite(GSEAres, file = paste0(filename, '_gsea_results.tsv'), sep = "\t", sep2 = c("", " ", ""))
sessionInfo()
Check my sessionInfo() here in case you have trouble reproducting my steps:
> sessionInfo()
R version 4.2.3 (2023-03-15 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 22621)
Matrix products: default
locale:
[1] LC_COLLATE=English_Ireland.utf8 LC_CTYPE=English_Ireland.utf8
[3] LC_MONETARY=English_Ireland.utf8 LC_NUMERIC=C
[5] LC_TIME=English_Ireland.utf8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] fgsea_1.24.0 RColorBrewer_1.1-3 lubridate_1.9.2 forcats_1.0.0
[5] stringr_1.5.0.9000 dplyr_1.1.2 purrr_1.0.1 readr_2.1.4
[9] tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.2 tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.11 pillar_1.9.0 compiler_4.2.3 tools_4.2.3
[5] lattice_0.20-45 lifecycle_1.0.3 gtable_0.3.3 timechange_0.2.0
[9] pkgconfig_2.0.3 rlang_1.1.1 Matrix_1.5-3 fastmatch_1.1-3
[13] cli_3.6.0 rstudioapi_0.14 parallel_4.2.3 withr_2.5.0
[17] generics_0.1.3 vctrs_0.6.3 hms_1.1.3 cowplot_1.1.1
[21] grid_4.2.3 tidyselect_1.2.0 data.table_1.14.8 glue_1.6.2
[25] R6_2.5.1 fansi_1.0.4 BiocParallel_1.32.6 farver_2.1.1
[29] tzdb_0.4.0 magrittr_2.0.3 codetools_0.2-19 scales_1.2.1
[33] colorspace_2.1-0 labeling_0.4.2 utf8_1.2.3 stringi_1.7.12
[37] munsell_0.5.0 And that is the end of this tutorial!
In this post, I explained how to perform gene set enrichment analysis using fgsea. Hope you found it useful!
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and… see you in the next one!
Squids don't care much for coffee,
but Laura loves a hot cup in the morning!
If you like my content, you might consider buying me a coffee.
You can also leave a comment or a 'like' in my posts or Youtube channel, knowing that they're helpful really motivates me to keep going:)
Cheers and have a 'squidtastic' day!


0 Comments