A short but simple explanation of PCA – easily explained with an example!
PCA, t-SNE, UMAP… you’ve probably heard about all these dimensionality reduction methods. In this series of blogposts, we’ll cover the similarities and differences between them, easily explained!
In this post, you will find out what is PCA and how to interpret it with an example.
So if you are ready… let’s dive in!
Click on the video to follow my easy PCA explanation with an example on Youtube!
What is dimensionality reduction?
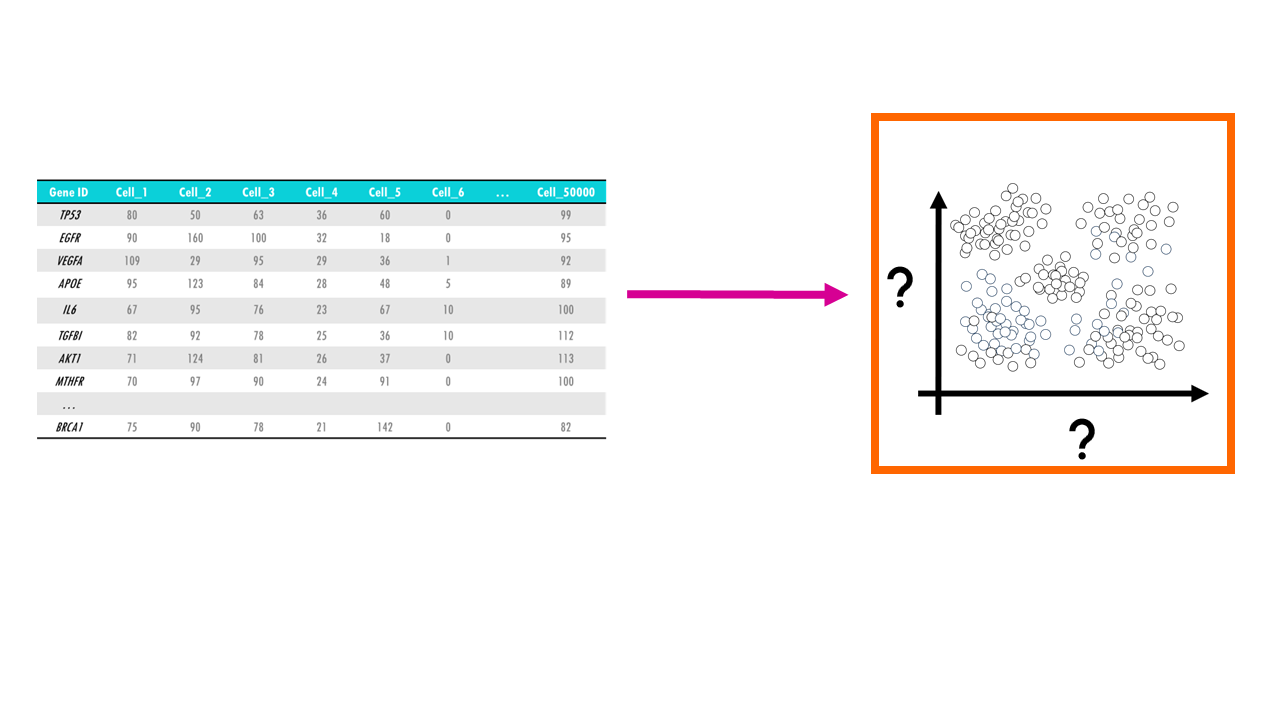
Let’s imagine you have a single-cell dataset with the gene expression of 10000 genes across 50.000 cells. For each cell, and for each gene, you have a gene expression value that tells you how much that gene was expressed in that cell.
Sometimes, we just want to visualise large datasets like this one in a plot. If each point in the plot is a cell, that would allow us to identify clusters of cells with similar expression levels of certain genes, we can identify cell types, or cells that are very different to the rest in their gene expression profile. The problem with such big datasets like this one, is that we cannot visualise all features, or genes, at once. A plot will have two axes, three at most. So we could pick two genes, and plot each cell according to their expression level of MCM3 and AKT1. For example, you might identify a cluster of cells which have a high expression of both MCM3 and AKT1, and a cluster of cells with a high expression of MCM3 but relatively low expression of AKT1. However, this plot leaves out the expression level of the other 9998 genes in our dataset.
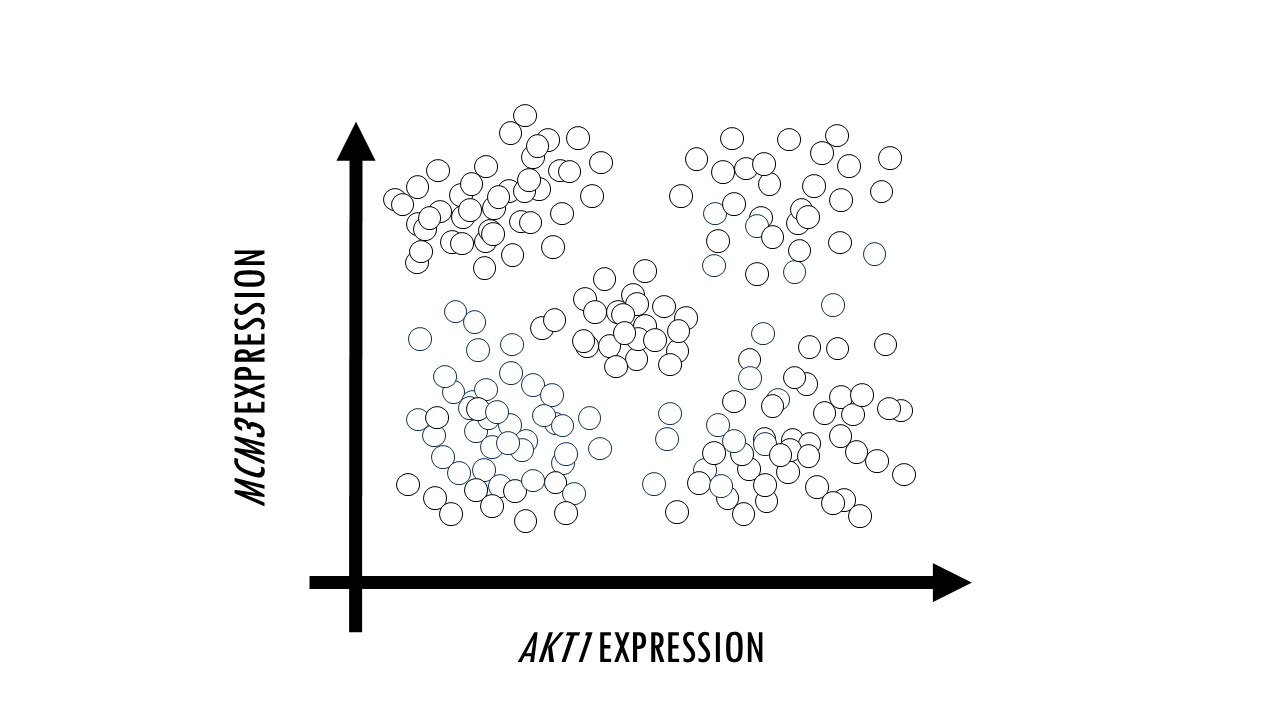
In other words, we need a way to convert a multidimensional dataset into two variables we can plot. This process is called dimensionality reduction and through it we basically reduce the number of input variables or features (in this case, genes) while retaining maximum information. Obviously, when we reduce the number of dimensions, we are going to lose information, but the idea is to preserve as much of the dataset’s structure and characteristics as possible.
To do this, we need a dimension reduction algorithm, which is basically a method that reduces the number of dimensions. As you can imagine, there are many dimension reduction algorithms out there, but we are going to talk about 3 of the most common ones: PCA, UMAP and t-SNE. We will go through the main ideas of each of them and the differences between them, so you can decide which one is most appropriate for your data.
Squidtip
Dimensionality reduction is a process that transforms data from high-dimensional space into a low-dimensional space while preserving as much of the dataset’s original information as possible.
Principal Component Analysis (PCA)
Let’s start with PCA or Principal Component Analysis. PCA is a linear dimension reduction algorithm. We’ll talk about linear vs non-linear later on in the series. As a dimension reduction algorithm, with PCA you basically transform your data from a high dimensional space (so many features) to a low-dimensional space.
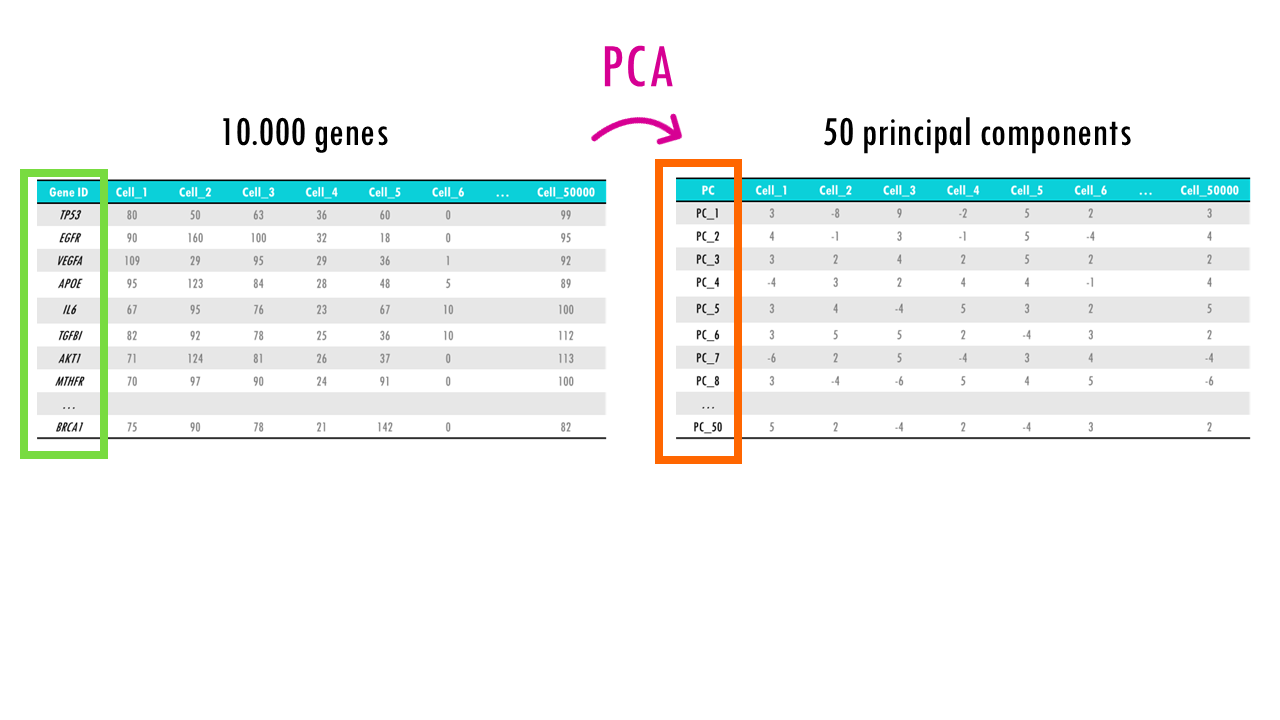
With PCA you basically end up with principal components or PCs. For each cell, we will have a score for PC1, PC2, PC3 etc… But what is a principal component?
In simple terms, a principal component (PC) is like a summary of the most important patterns or variations in your data. For each PC, all the genes of the dataset have a score or loading, which can be negative or positive and have a bigger or smaller value, depending on if they contribute more or less to that principal component.
If we go back to our single-cell RNAseq dataset, we have a lot of genes in our dataset. But not all genes contribute equally to the differences between cells. Some genes might be more informative or change more across the cells, while others are less important.
Principal components are new “axes” (directions) in the data that capture the biggest differences between cells. These directions are chosen so that:
- The first principal component (PC1) captures the largest possible variation across all cells. This is the axis where the differences between cells are the greatest.
- The second principal component (PC2) captures the second-largest variation, but it is orthogonal (independent) to the first one.
- And so on for higher PCs.
Squidtip
Principal Components are ranked and orthogonal (independent from each other): the first principal component (PC1) captures the largest possible variation across all cells. PC2 captures the second largest variation across all cells, that was not captured by PC1…, and so on.
Let’s check the PCA results for our dataset. We know that PC1 is the most important principal component and holds the most variability in our dataset, so let’s have a closer look at what it represents. Remember that each PC has scores for all of the genes. We notice that genes involved in cell division and promote cell cycle have very high positive values for PC1, whereas genes that code for negative regulators of cell division and are related to cell senescence or cell death have very high negative values. Genes involved, in let’s say, immune function, have values very close to 0. In a way, PC1 has summarised all genes in a sort of “meta” feature representing cell division or proliferation. So if we plot all cells by their PC1 score we know that cells with high PC1 score are generally cells that are dividing, whilst cells that have very low scores for PC1 are dying or senescent. Just plotting PC1 will not let us differentiate between healthy cells like epithelial cells or B cells that are dividing a lot, and cancer cells which also proliferate a lot. But it’s a start.
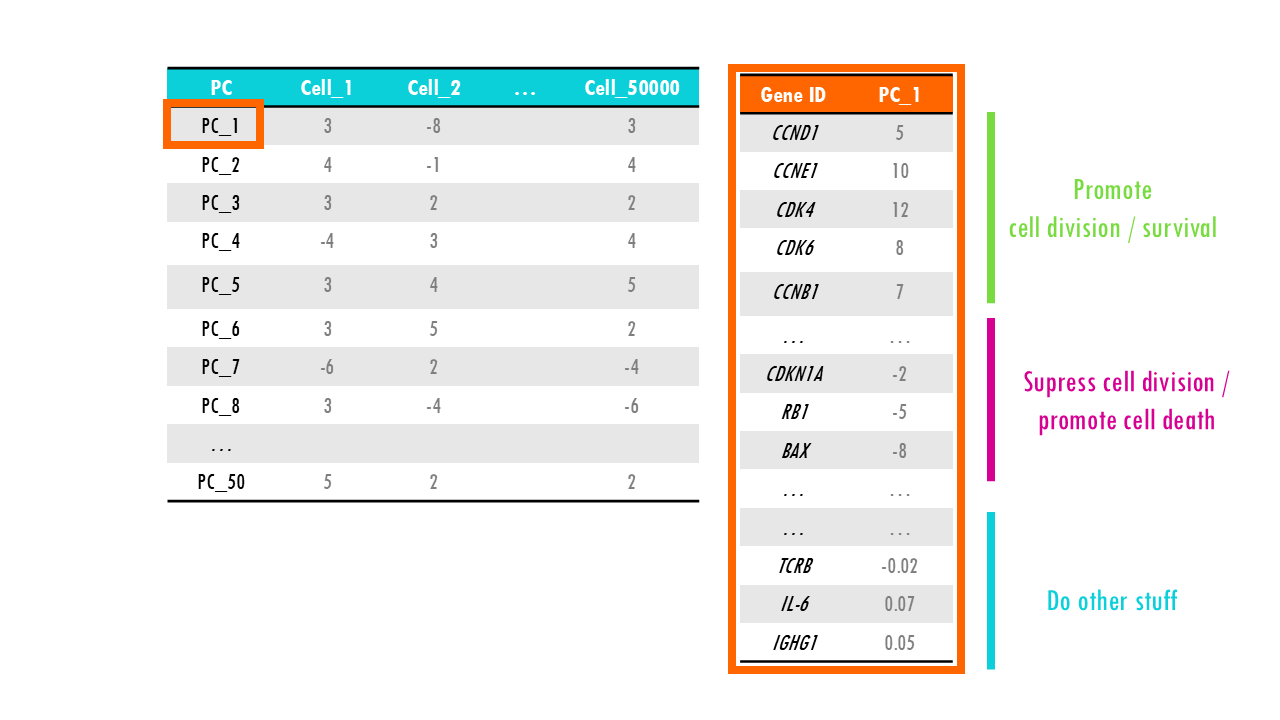
We need more PCs, so let’s have a look at the next principal component which holds the next largest information, or variation of our dataset. We find that PC2 represents “immune function”; so cells with very high PC2 scores have a high expression of gene sets involved in the immune response. This way, if we now add an axis to our plot, we can visualise cells that divide a lot and are involved in the immune response, so maybe B cells or monocytes, immune cells that are not dividing, for example, anergic Plasma cells or macrophages, cells that divide but are not immune cells, like skin cells, cancer cells or progenitor cells, and non-immune cells that do not divide at all, such as enterocytes or other differentiated cells.
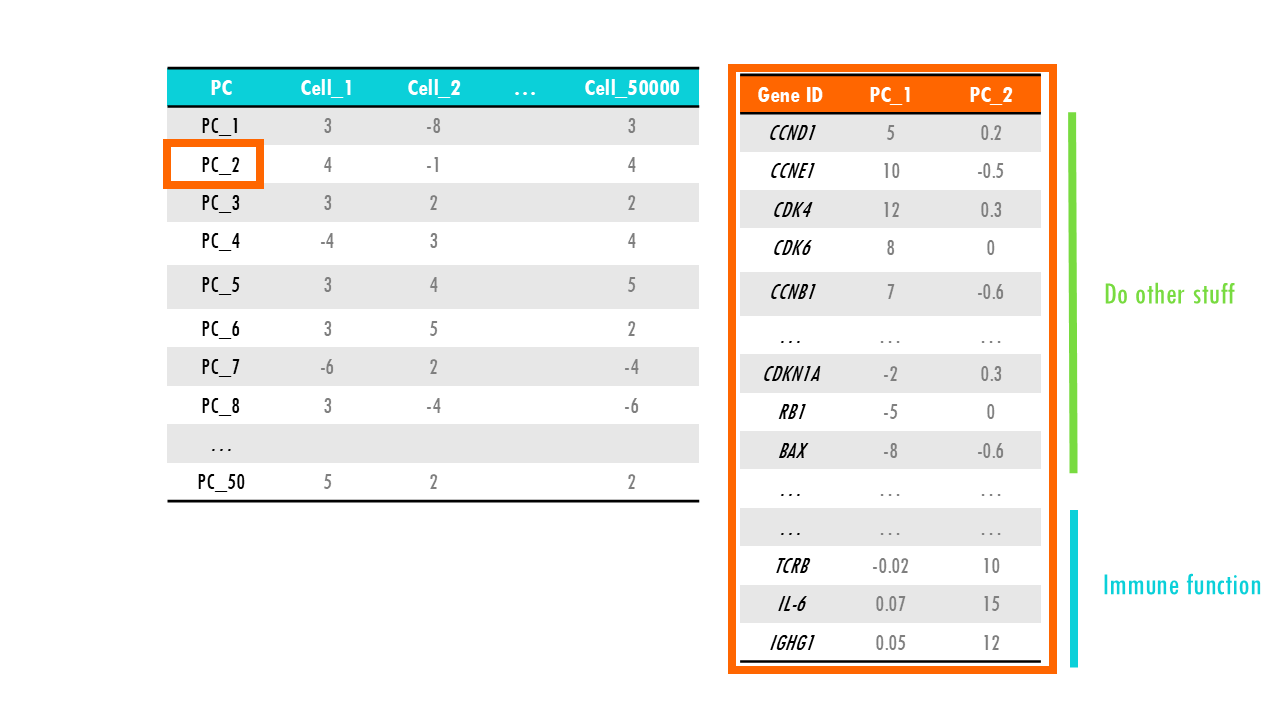
In real datasets it is not often as simple as this example, but in essence, PCA summarises gene expression profiles into a few principal components, making us understand the key patterns in the data without needing to look at every individual gene.
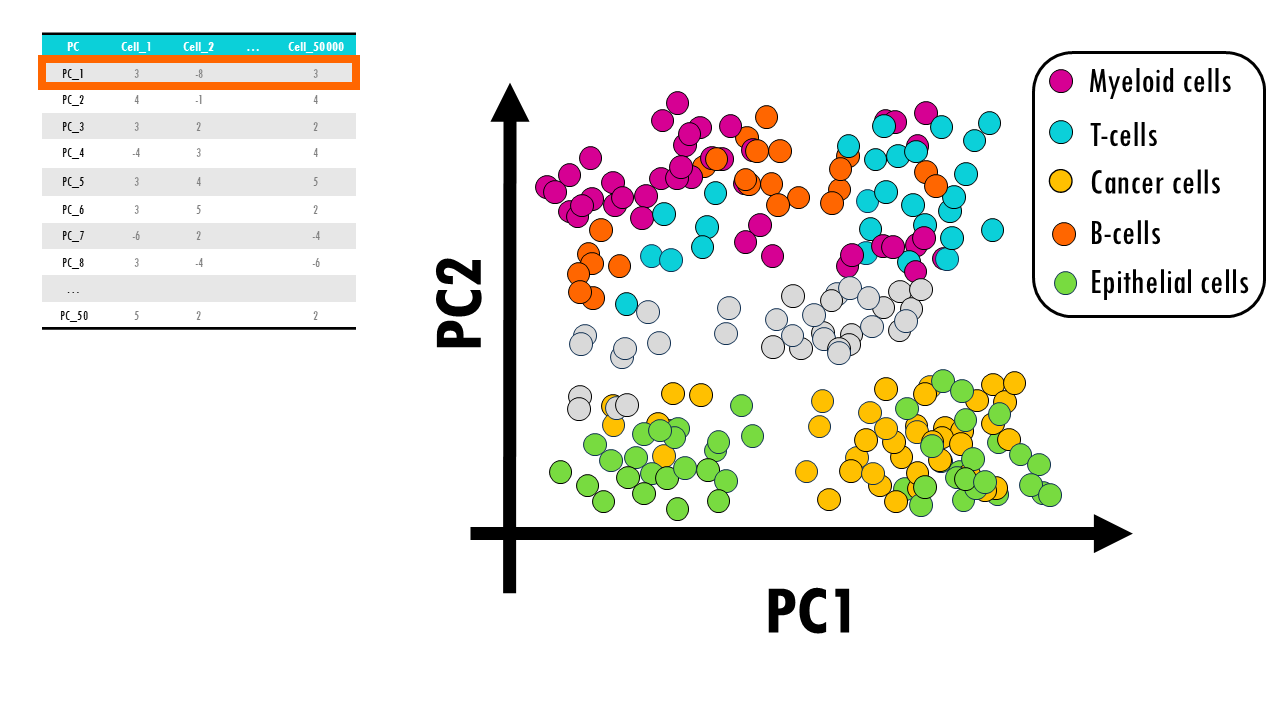
Ok, but you might think, great, we’re visualising cells that divide or have immune function or not, but what if we want to distinguish B cells from T cells? Or dividing, non immune cells like cancer cells from epithelial cells? Or different subtypes of epithelial cells?
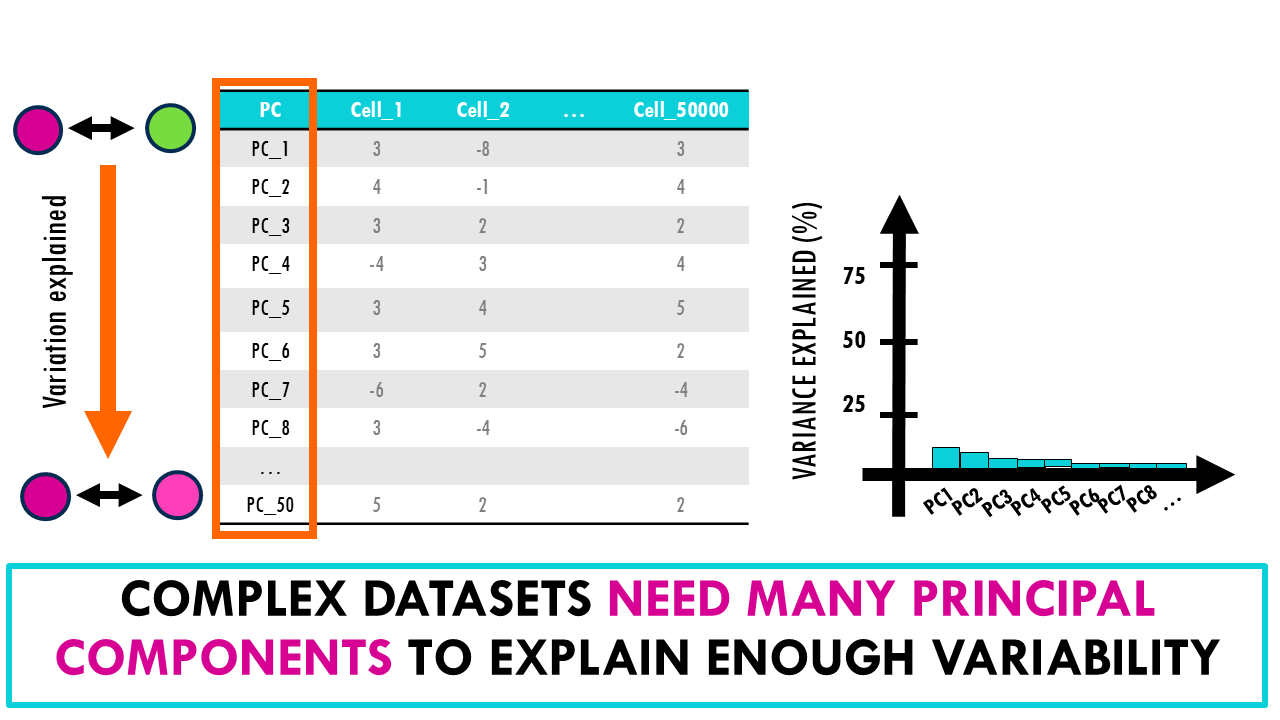
For that, we have to keep looking at different principal components. Since they are orthogonal, or independent, as we include more PCs, we take into account more sources of variation in our dataset, making us able to distinguish more fine differences between our cells. Often, we compute 30, 50 or 100 PCs. A scree plot can help us see how much of the variability in the dataset does each PC hold. If the first two PCs hold most of the variability, then great! You can get a pretty good overview of your entire dataset by just plotting the first two PCs.
However, often in biological datasets the scree plot looks like this, where the variability of the dataset is explained by 10, 20 or more principal components. This is too many dimensions to plot. We need a different dimensionality reduction algorithm.
Squidtip
A scree plot helps us see how much of the total variability in the dataset is explained by each PC.
Final notes on PCA
In summary, PCA is a dimensionality reduction algorithm which allows us to summarise many thousand features (e.g., genes) into a few dozen Principal Components or PCs. These PCs are ranked and orthogonal, meaning the first PC holds the most variability in the dataset, then the second and so on.
If you’d like a more thorough explanation on how PCA works, you can check out my other blogpost: PCA easily explained.
Otherwise, keep reading part 2 of this series: where we’ll talk about another great dimensionality reductio algorithm: t-SNE!
Ending notes
Wohoo! You made it ’til the end!
In this post, I shared some insights on Principal Component Analysis and how to interpret a PCA plot.
Hopefully you found some of my notes and resources useful! Don’t hesitate to leave a comment if there is anything unclear, that you would like explained further, or if you’re looking for more resources on biostatistics! Your feedback is really appreciated and it helps me create more useful content:)
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and… see you in the next one!
Squids don't care much for coffee,
but Laura loves a hot cup in the morning!
If you like my content, you might consider buying me a coffee.
You can also leave a comment or a 'like' in my posts or Youtube channel, knowing that they're helpful really motivates me to keep going:)
Cheers and have a 'squidtastic' day!
And that is the end of this tutorial!
In this post, I explained the differences between log2FC and p-value, and why in differential gene expression analysis we don't always get both high log2FC and low p-value. Hope you found it useful!
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and... see you in the next one!


I see an abundance of Youtube.com sites trying to explain a variety of tests and mechanisms of cell biology, and this time I was looking for PCA (principal component analysis) and came across biostatsquid.com. I think this youtube on PCA was exceptional, great pace and clarity of concepts and marvelous animated illustrations, that are finally complemented with an explanatory text which makes the site once of a kind. Thank you and congratulations.
Thank you so, so much! I’m really happy it helped you and so glad you enjoyed it too!