A step-by-step R tutorial on performing pathway enrichment analysis with clusterProfiler in R
Note! This is an updated version of ORA analysis with clusterProfiler which doesn’t correspond exactly to the video tutorial! Click here for the old version.
Table of contents
Before you start
Step-by-step clusterProfiler tutorial
Step 0: Set up your script and load the data
Step 1: Prepare inputs for clusterProfiler
Step 2: Run clusterProfiler
Step 3: Visualise your pathway enrichment analysis results
SessionInfo()
Before you start…
Welcome to Biostatsquid’s easy and step-by-step tutorial on ClusterProfiler! In this guide, we will explore an essential tool for functional enrichment analysis and interpretation of gene sets or clusters of genes.
So what does clusterProfiler do? Functional enrichment analysis plays a crucial role in understanding the biological processes, molecular functions, and cellular components associated with a set of genes. CluserProfiler will allow us to identify and visualize enriched functional terms, such as Gene Ontology (GO) terms and biological pathways, within our gene sets.
Throughout this tutorial, we will walk you through the key features and steps involved in using ClusterProfiler. You will learn how to annotate genes, perform enrichment analysis, apply multiple testing correction, and interpret the results. In part 2, we will explore the various visualization techniques available in ClusterProfiler, helping you gain a comprehensive understanding of the enriched functional annotations.
For this tutorial you will need R, or Rstudio, and you will need to install the following packages:
# For data management
install.packages('tidyverse')
BiocManager::install("clusterProfiler")
BiocManager::install("org.Hs.eg.db")
# For visualisation
install.packages('pheatmap')
install.packages("DOSE")
install.packages("enrichplot")
install.packages("ggupset")If you are a beginner in R, don’t be overwhelmed! This tutorial will go step-by-step and I will explain (almost!) every line of code so you know what is happening at each point of the workflow.
You might also want to check out my Youtube tutorial on how to do pathway enrichment analysis using clusterProfiler. Or if you prefer a written step-by-step guide, then continue reading and don’t forget you can just copy the code by clicking on the top right corner of the code snippets in this post.
But enough introduction! Ready to enter the world of functional enrichment analysis with ClusterProfiler? Let’s dive in!
Squidtip
Want to know more about functional enrichment analysis? Check out my blogposts on Pathway Enrichment Analysis and Gene Set Enrichment Analysis – simply explained!
In this Youtube video, I explain how to do Pathway Enrichment Analysis with the R package clusterProfiler()!
Step-by-step clusterProfiler tutorial
The dataset
For this tutorial, I will use the differential gene expression results we used in my volcano plots tutorial. It is the human COVID T cell single-cell RNA-seq dataset from Bacher et al. (2020). You can read more about the dataset and obtain it for free here.
I already pre-processed the data and carried out the differential gene expression analysis between severe and healthy samples for Tfh-like cells. To follow this tutorial, you can just download the DGE results here. Right click, then ‘Save link as’.
If you would like to use your own data, you just need a simple gene expression dataframe with the following columns:
- gene_symbol: the gene symbols (or IDs, i.e., ENSEMBLE IDs)
- pval: p-values from your differential gene expression analysis.
- padj: p-adjusted values from your differential gene expression analysis (p-value is enough)
- log2fc: your log2 fold change values, showing the direction of the change (upregulation or downregulation).
Step 0: Set up your script and load the data
Before we start, it’s always good practice to clean-up our environment in case we have hidden objects in the background, free up memory, and set our working directory. We also need to load the necessary libraries. Today we will be working with clusterProfiler() package, and several other R libraries for data visualisation and management. You might need to install them first using install.packages() or BiocManager::install().
# Setting up environment ===================================================
# Clean environment
rm(list = ls(all.names = TRUE)) # will clear all objects including hidden objects
gc() # free up memory and report the memory usage
options(max.print = .Machine$integer.max, scipen = 999, stringsAsFactors = F, dplyr.summarise.inform = F) # avoid truncated output in R console and scientific notation
# Loading relevant libraries
library(tidyverse) # includes ggplot2, for data visualisation. dplyr, for data manipulation.
library(RColorBrewer) # for a colourful plot
library(pheatmap)
library(clusterProfiler) # for PEA analysis
library(enrichplot) # for visualisations
library(ggupset) # for visualisations
## Set relevant paths
list.files()
in_path <- "C:/Users/laura/Documents/Biostatsquid/Projects/data/scRNAseq"
out_path <- "C:/Users/laura/Documents/Biostatsquid/Projects/PEA/clusterProfiler"
dir.create(out_path, showWarnings = F, recursive = T)
Let’s get started! The first step is of course to load our differential gene expression results. If you have your own dataset, just read it into RStudio, and make sure you have a dataframe as I describe above. We can also add a new column, diffexpressed, which indicates whether a gene is UPregulated, DOWNregulated or NOt differentially expressed. I used a log2fc = 0 and p-adjusted value of 0.05 to define the thresholds, but you might want to tweak it depending on how permissive you want to be (or how many differentially expressed genes you have!).
# Read in data ===================================================
list.files(in_path)
df <- read.csv(paste0(in_path, 'severevshealthy_degresults.csv'), row.names = 1)
# Annotate according to differential expression
df <- df %>% mutate(diffexpressed = case_when(
log2fc > 0 & padj < 0.05 ~ 'UP',
log2fc < 0 & padj < 0.05 ~ 'DOWN',
padj > 0.05 ~ 'NO'
))Squidtip
Did you notice? Instead of writing the full path of my data file (e.g., ‘C:/Users/laura/Documents/Biostatsquid/Projects/data/scRNAseq/severevshealthy_degresults.csv’), I just pasted together the variable in_path (which I previously defined), and the name of the file, ‘severevshealthy_degresults.csv’ with the file.path() function.
Ok! So before we can start our analysis we need to check what clusterProfiler needs. If you would like to read more about the method, I recommend reading the publication. The reference manual lists all the functions of the package.
For the sake of making this tutorial as generalisable as possible, I will explain how to use the enricher() function which allows you to use any set of genes (also known as pathways).
- We will use the R package msigdbr to access our gene sets. You ccan read more about gene pathways here.
- However, clusterProfiler already provides functions for some pathway databases like enrichKEGG() and enrichGO(). So take a look at the clusterProfiler package, it has a lot to offer!
Step 1: Prepare inputs for clusterProfiler
Prepare pathways or gene sets
1. Get your gene set .gmt files. In my old version (and in the video), I used to download the .gmt files with the pathways manually from the msigdb database. Now, it’s so much easier! You can use the R package msigdbr to download whichever pathways you’re interested in. If you are unsure which one is right for you, check out this other post where I explain the different pathways and gene sets available in msigdb.
Make sure to download the gene sets that are appropriate for your analysis, and edit the species if needed!
## Prepare pathways or gene sets -----------------------------------------------
# msigdbr R package provides Molecular Signatures Database (MSigDB) gene sets
# Read more here: https://igordot.github.io/msigdbr/articles/msigdbr-intro.html
# msigdbr_collections()
name_of_gene_set <- 'H' # for hallmark gene sets
# Other options:
# 'C1' - positional gene sets
# 'C2' - curated gene sets (includes KEGG, Reactome, BioCarta, etc.)
# 'C3' - regulatory target gene sets
# 'C4' - computational gene sets
# 'C5' - ontology gene sets (GO terms)
# 'C6' - oncogenic signature gene sets
# 'C7' - immunologic signature gene sets
# 'C8' - cell type signature gene sets
gene_sets_df <- msigdbr(species = 'Homo sapiens', category = name_of_gene_set)
#gene_sets_df <- msigdbr(species = 'Homo sapiens', category = 'C5', subcollection = 'GO:BP')
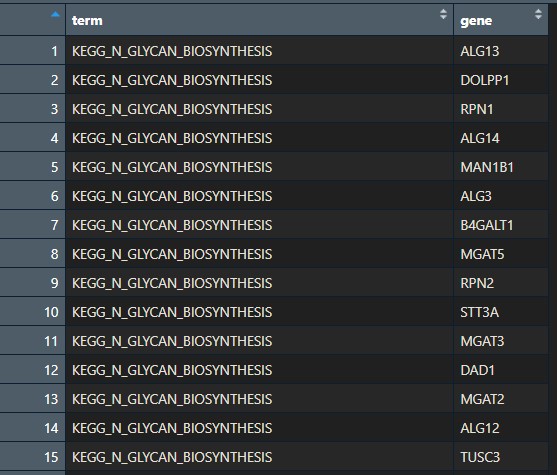
head(gene_sets_df)2. Filter the .gmt file and convert it to a format accepted by clusterProfiler: the first column should have pathway names, the second column should have gene symbols. You can use the following code:
# Convert to named list format required by clusterprofiler: the first column should have pathway names
# the second column should have gene symbols
gene_sets <- gene_sets_df %>%
dplyr::select(gs_name, gene_symbol)
cat(paste("Loaded", length(unique(gene_sets$gs_name)), "gene sets\n"))Basically, it should look like this:
Prepare differential gene expression results
We will:
- Remove non-significant genes: since we only want the enriched pathways of the significantly differentially expressed genes.
- Substitute names so they are annotated nicely in the heatmap later
- Split the data into a list of 2 sub-dataframes: upregulated, downregulated genes.
## 2. Prepare deg data -----------------------------------------------
# Annotate according to differential expression
df <- df %>% mutate(diffexpressed = case_when(
log2fc > 0 & padj < 0.05 ~ 'UP',
log2fc < 0 & padj < 0.05 ~ 'DOWN',
padj > 0.05 ~ 'NO'
))
# Remove non-significant genes for ORA
df_sig <- df[df$diffexpressed != 'NO', ] # remove non-significant genes
cat(paste("Found", nrow(df_sig), "significantly differentially expressed genes\n"))
cat(paste("UP:", sum(df_sig$diffexpressed == 'UP'), "\n"))
cat(paste("DOWN:", sum(df_sig$diffexpressed == 'DOWN'), "\n"))
# Split the dataframe into a list of sub-dataframes: upregulated, downregulated genes
deg_results_list <- split(df_sig, df_sig$diffexpressed)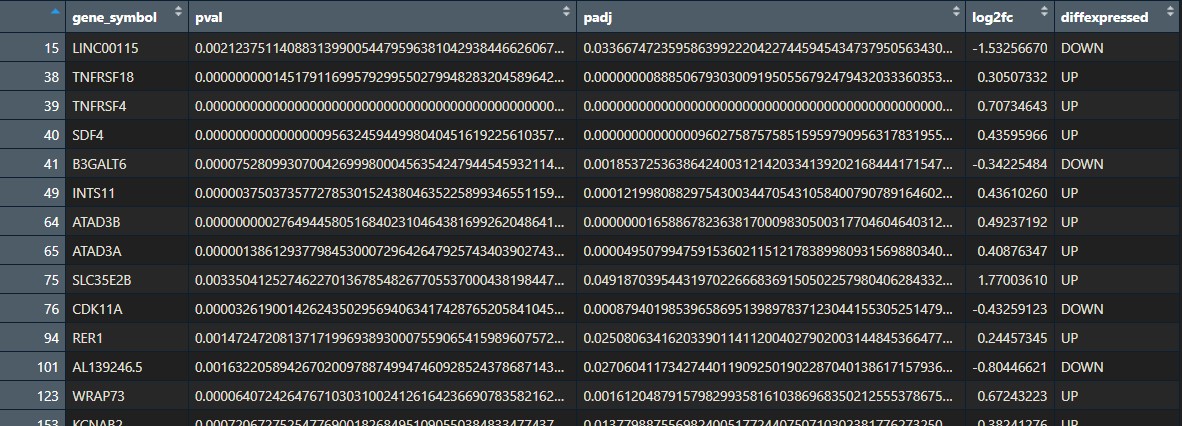
Prepare background genes
As I mentioned in my previous blogpost, it’s important to provide a set of background genes against which to compare your DGE results. This helps avoid bias and get more accurate statistical results.
For example, if you are studying neural cells, it doesn’t make sense to include muscle-specific genes when deciding whether a pathway is upregulated or not – those genes could have never been expressed in neural cells so they shouldn’t be taken into account.
Sometimes it’s hard to decide what “could have been” measured in your experiment, but a good approximation is to include all genes you detected as part of your “gene universe”.
## 4. Prepare background genes ----------------------------------------
# These should be ALL the genes you could have measured in your experiment,
# to avoid bias. I will use all the genes from the original dataset we used
# to compare severe vs healthy cells
data <- scRNAseq::BacherTCellData(filtered = TRUE, ensembl = FALSE, location = TRUE)
background_genes <- row.names(data)
background_genes[1:5]
rm(data) # to clear some spaceStep 2: Run clusterProfiler
Wohoo! After all these steps… we are finally ready to run clusterProfiler! Are you excited?
First, let’s set some variables. This is not essential but it will make your life easier in case you need to change them later on.
## 3. Run ClusterProfiler -----------------------------------------------
# Settings
name_of_comparison <- 'severevshealthy'
padj_cutoff <- 0.05
genecount_cutoff <- 5
filename_prefix <- paste0(name_of_comparison, '_', name_of_gene_set)Squidtip
You don’t need to set all these variables to run clusterProfiler.
But! If you have a lot of differential gene expression results and/or gene sets… you will have to copy paste your code maaany times and just change a few names… This is very tedious, time-consuming and error-prone! A bit of code can save you a lot of time. If you haven’t thought about it yet- I’m talking about building your own function!
These variables set-up is a good place to start. If you are interested in a blogpost explaining how to create a function that runs clusterProfiler and outputs your results and plots by just giving it a few variables – leave me a comment down below!
Now that we have our pathways and our differential expressed genes ready… let’s call the enricher() function from clusterProfiler.
We use lapply as we are applying the function to our list (remember we split the data into a list of 2 subsets).
Squidtip
If you only have 1 dataset (e.g., just upregulated genes results), you can run the enricher function directly:
x <- enricher(my_dge_results, TERM2GENE = my_pathways, universe = my_background_genes)# Run clusterProfiler on each sub-dataframe
res <- lapply(names(deg_results_list),
function(x) enricher(gene = deg_results_list[[x]]$gene_symbol,
TERM2GENE = gene_sets,
universe = background_genes,
# Let's use the cut-offs later to filter ORA results
#pvalueCutoff = padj_cutoff,
#minGSSize = genecount_cutoff,
qvalueCutoff = 0.2))
names(res) <- names(deg_results_list)
# Check results
cat("Enrichment analysis results:\n")
for(i in names(res)) {
cat(paste(i, ":", nrow(as.data.frame(res[[i]])), "enriched pathways\n"))
}Squidtip
What does the enricher() function do?
It basically does a hypergeometric test for pathway enrichment analysis, accepting a user-defined annotation. So it is very useful if you want to analyse your data with unsupported organisms or other ontologies/pathways, or customised annotations.
Its inputs are the dge results, and two additional parameters TERM2GENE and TERM2NAME.
TERM2GENEis a data.frame with first column of term ID and second column of corresponding mapped gene. It is used to match the pathway with the genes.TERM2NAMEis optional, used to rename pathways. It is adata.framewith first column of term ID and second column of corresponding term name.
More information here: https://github.com/YuLab-SMU/clusterProfiler
Great! We now have a list with our pathway enrichment analysis results. Let’s make it more readable and convert it to a dataframe with the following lines of code:
#Convert the enrichResults to a dataframe with the pathways
res_df <- lapply(names(res), function(x) rbind(res[[x]]@result))
names(res_df) <- names(res)
res_df <- do.call(rbind, res_df)
head(res_df)And maybe add a few more columns to our dataframe ):
res_df_sig <- res_df_sig %>% mutate(minuslog10padj = -log10(p.adjust))Now, we can subset our results. Filter the significant pathways according to p-adjusted threshold and gene count.
# Subset to those pathways that have p adj < cutoff and gene count > cutoff (you can also do this in the enricher function)
res_df_sig <- res_df_sig %>%
dplyr::filter(p.adjust < padj_cutoff & Count > genecount_cutoff) # select only target pathways have p adjusted < 0.05 and at least 6 genes
Squidtip
Feel free to adjust these thresholds to your own results! You might need to play around with the numbers until you find what best suits your dataset and your goal.
Finally, don’t forget to save your results!
print('Saving clusterprofiler results')
write.csv(res_df_sig, file.path(out_path, paste0(filename_prefix, '_resclusterp.csv')), row.names = FALSE)
# Also save the enrichresults object for plotting later
qs::qsave(res, file.path(out_path, paste0(filename_prefix, '_resclusterp_enrichres.qs')))
# You can also save it as RDS
Step 3: Visualise your pathway enrichment analysis results
Now it’s time for the fun part! Let’s explore our results by plotting them in different ways.
Check how to visualise your clusterProfiler results and create pretty plots here: Visualisation of pathway enrichment results.
sessionInfo()
Check my sessionInfo() here in case you have trouble reproducting my steps:
R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 26100)
Matrix products: default
locale:
[1] LC_COLLATE=English_Ireland.utf8 LC_CTYPE=English_Ireland.utf8 LC_MONETARY=English_Ireland.utf8 LC_NUMERIC=C
[5] LC_TIME=English_Ireland.utf8
time zone: Europe/Dublin
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] enrichplot_1.26.6 DOSE_4.0.1 clusterProfiler_4.14.6 msigdbr_24.1.0 ggupset_0.4.1
[6] RColorBrewer_1.1-3 lubridate_1.9.4 forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4
[11] purrr_1.0.4 readr_2.1.5 tidyr_1.3.1 tibble_3.2.1 ggplot2_3.5.1
[16] tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] ProtGenerics_1.38.0 bitops_1.0-9 fs_1.6.5 matrixStats_1.5.0
[5] spatstat.sparse_3.1-0 httr_1.4.7 alabaster.base_1.6.1 tools_4.4.2
[9] sctransform_0.4.1 utf8_1.2.4 R6_2.6.1 HDF5Array_1.34.0
[13] lazyeval_0.2.2 uwot_0.2.3 rhdf5filters_1.18.1 withr_3.0.2
[17] sp_2.2-0 gridExtra_2.3 progressr_0.15.1 cli_3.6.4
[21] Biobase_2.66.0 spatstat.explore_3.4-2 fastDummies_1.7.5 alabaster.se_1.6.0
[25] labeling_0.4.3 Seurat_5.2.1 spatstat.data_3.1-6 ggridges_0.5.6
[29] pbapply_1.7-2 Rsamtools_2.22.0 yulab.utils_0.2.0 gson_0.1.0
[33] R.utils_2.13.0 parallelly_1.42.0 limma_3.62.2 rstudioapi_0.17.1
[37] RSQLite_2.4.1 BiocIO_1.16.0 generics_0.1.3 gridGraphics_0.5-1
[41] RApiSerialize_0.1.4 ica_1.0-3 spatstat.random_3.3-3 GO.db_3.20.0
[45] Matrix_1.7-1 S4Vectors_0.44.0 abind_1.4-8 R.methodsS3_1.8.2
[49] lifecycle_1.0.4 yaml_2.3.10 edgeR_4.4.2 SummarizedExperiment_1.36.0
[53] rhdf5_2.50.2 qvalue_2.38.0 SparseArray_1.6.2 BiocFileCache_2.14.0
[57] Rtsne_0.17 grid_4.4.2 blob_1.2.4 promises_1.3.2
[61] ExperimentHub_2.14.0 crayon_1.5.3 miniUI_0.1.1.1 speckle_1.6.0
[65] ggtangle_0.0.7 lattice_0.22-6 cowplot_1.1.3 GenomicFeatures_1.58.0
[69] KEGGREST_1.46.0 pillar_1.10.1 fgsea_1.32.4 GenomicRanges_1.58.0
[73] rjson_0.2.23 future.apply_1.11.3 codetools_0.2-20 fastmatch_1.1-6
[77] glue_1.8.0 ggfun_0.2.0 spatstat.univar_3.1-2 data.table_1.17.0
[81] gypsum_1.2.0 vctrs_0.6.5 png_0.1-8 treeio_1.30.0
[85] spam_2.11-1 gtable_0.3.6 assertthat_0.2.1 cachem_1.1.0
[89] S4Arrays_1.6.0 mime_0.12 survival_3.7-0 SingleCellExperiment_1.28.1
[93] statmod_1.5.0 fitdistrplus_1.2-2 ROCR_1.0-11 nlme_3.1-166
[97] ggtree_3.14.0 bit64_4.6.0-1 alabaster.ranges_1.6.0 filelock_1.0.3
[101] RcppAnnoy_0.0.22 GenomeInfoDb_1.42.3 irlba_2.3.5.1 KernSmooth_2.23-24
[105] colorspace_2.1-1 BiocGenerics_0.52.0 DBI_1.2.3 tidyselect_1.2.1
[109] bit_4.6.0 compiler_4.4.2 curl_6.2.1 httr2_1.1.0
[113] scRNAseq_2.20.0 DelayedArray_0.32.0 plotly_4.10.4 rtracklayer_1.66.0
[117] stringfish_0.16.0 scales_1.3.0 lmtest_0.9-40 rappdirs_0.3.3
[121] digest_0.6.37 goftest_1.2-3 spatstat.utils_3.1-3 alabaster.matrix_1.6.1
[125] XVector_0.46.0 htmltools_0.5.8.1 pkgconfig_2.0.3 MatrixGenerics_1.18.1
[129] ensembldb_2.30.0 dbplyr_2.5.0 fastmap_1.2.0 rlang_1.1.5
[133] htmlwidgets_1.6.4 UCSC.utils_1.2.0 shiny_1.10.0 farver_2.1.2
[137] zoo_1.8-13 jsonlite_1.9.0 BiocParallel_1.40.2 GOSemSim_2.32.0
[141] R.oo_1.27.1 RCurl_1.98-1.17 magrittr_2.0.3 GenomeInfoDbData_1.2.13
[145] ggplotify_0.1.2 dotCall64_1.2 patchwork_1.3.0 Rhdf5lib_1.28.0
[149] munsell_0.5.1 Rcpp_1.0.14 ape_5.8-1 babelgene_22.9
[153] ggnewscale_0.5.2 reticulate_1.41.0.1 alabaster.schemas_1.6.0 stringi_1.8.4
[157] zlibbioc_1.52.0 MASS_7.3-61 AnnotationHub_3.14.0 plyr_1.8.9
[161] parallel_4.4.2 listenv_0.9.1 ggrepel_0.9.6 deldir_2.0-4
[165] Biostrings_2.74.1 splines_4.4.2 tensor_1.5 hms_1.1.3
[169] locfit_1.5-9.12 igraph_2.1.4 spatstat.geom_3.3-6 RcppHNSW_0.6.0
[173] reshape2_1.4.4 stats4_4.4.2 XML_3.99-0.18 BiocVersion_3.20.0
[177] SeuratObject_5.0.2 RcppParallel_5.1.10 renv_1.0.9 BiocManager_1.30.25
[181] tzdb_0.5.0 httpuv_1.6.15 RANN_2.6.2 polyclip_1.10-7
[185] alabaster.sce_1.6.0 qs_0.27.3 future_1.34.0 scattermore_1.2
[189] xtable_1.8-4 AnnotationFilter_1.30.0 restfulr_0.0.16 RSpectra_0.16-2
[193] tidytree_0.4.6 gitcreds_0.1.2 later_1.4.1 viridisLite_0.4.2
[197] aplot_0.2.8 GenomicAlignments_1.42.0 memoise_2.0.1 AnnotationDbi_1.68.0
[201] IRanges_2.40.1 cluster_2.1.6 timechange_0.3.0 globals_0.16.3 And that is the end of this tutorial!
In this post, I explained how to perform functional enrichment analysis using clusterProfiler. Hope you found it useful!
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and… see you in the next one!


This is a really helpful to us. Thank you very much for this great effort.
We are also expecting ROC, OPLDA and PCA.
Hi Mukhtar, thank you so much for your comment and suggestions! Definitely great ideas for my upcoming posts/videos:) I already have a post on PCA which you can find here: https://biostatsquid.com/pca-simply-explained/ (but if there is something particular you would like explained about PCA, do let me know!)
very Informative,🌹🌹 waiting for visualization🥺
Hi Muddasar, thanks for your comment. You can find part 2, visualisation of pathway enrichment analysis results, here: https://biostatsquid.com/pathway-enrichment-analysis-plots/
Hi,
thank you for uploading this! it is so helpfull.
I have a question about this section:
bg_path <- "PEA/Background_genes/" # folder with your background genes used for PEA
this is a folder with background genes. but how do you create such folder? What type of file or directory should be uploaded in this folder?
Hi Raisa, thanks for your comment! bg_path is just a folder where I saved all the .gmt files which I downloaded from msigdb (check the section: ‘Prepare background genes’). You don’t really need it, you can also just save those files in your results folder and just use 1 folder for everything.
As for your question, how do you create it, you can just create it like any other folder, wherever you want it (I don’t know if you have Windows or Mac, but just on the file explorer > create folder > give it a name > and then just move your .gmt files there). Alternatively you can also create that folder within R with the command ‘dir.create()’.
If you check step 2 ‘2. Filter the .gmt file and convert it to a format accepted by clusterProfiler.’, you will see that when reading the .gmt file I am specifying the full path (file <- "PEA/Background_genes/c2.cp.kegg.v7.5.1.symbols.gmt") but you could also write: file <- paste0(bg_path, 'c2.cp.kegg.v7.5.1.symbols.gmt') Hopefully that made sense! If you have more questions, don't hesitate to ask:)
Hi there! Many many thanks for this tutorial…! I had a small question-
We filtered our DEGs against KEGG in the following step-
# Read in the .gmt file
file <- "PEA/Background_genes/c2.cp.kegg.v7.5.1.symbols.gmt"
pwl2 <- read.gmt(file)
# Subset to the genes that are present in our dataset
pwl2 <- pwl2[pwl2$gene %in% genes_in_data,]
# Save the filtered background gene set
filename <- 'kegg.RDS'
Then why did we choose REACTOME as bg_genes while running clusterProfiler.
# Settings
name_of_comparison <- 'severevshealthy' # for our filename
background_genes <- 'reactome' # for our filename
bg_genes <- readRDS(paste0(bg_path, 'reactome.RDS')) # read in the background genes
Shouldn't the later one be KEGG too with all genes (unfiltered)?
Apologies if I missed something or if asking a naive question! 🙂
Many many thanks:)
Hi! Thanks so much for your comment and the feedback:) You are totally right, well spotted!! I totally missed that one. Since I ran it with different gene sets I created all the .RDS files of filtered gene sets and then tested them out – I corrected it now just to keep it consistent. Thanks so much for letting me know!
Should the codes in this chunk:
# Subset to those pathways that have p adj cutoff (you can also do this in the enricher function)
target_pws <- unique(res_df$ID[res_df$p.adjust genecount_cutoff]) # select only target pathways have p adjusted < 0.05 and at least 6 genes
res_df <- res_df[res_df$ID %in% target_pws, ]
be corrected to:
res_df % filter(p.adjust genecount_cutoff)
as there are some cases when one of the two directions (up or down) of pathways with the same name do not pass padj_cutoff, so directly filtering the values themselves would be more accurate?
**res_df %>% filter(p.adjust genecount_cutoff)
Hi! Thanks so much for your comment, and sorry for the late reply.
I’m not sure if I understand – you’d like to filter only based on gene count? What I do is select only pathways that are significant (p-adj < 0.05) and have at least 6 genes. I think the code in your comment might be missing a few symbols - could you let me know again how you propose to filter the pathways? Since you already filtered for upregulated genes BEFORE you ran clusterProfiler, the results should only show you pathways that are enriched with up- OR downregulated genes. But please let me know what you mean! And thanks so much for your improvement suggestion - always happy to correct my code:)
Hi there!
I’m newbie to R, so pretty sure I’m missing something obvious.
When I read a .gmt file (KEGG for example), pwl2 shows 12797 obs and 2 variables
plus, genes_in_data from my deg list return 14695 elements. So far, so good.
Now, to check the subset to the genes that are present in my dataset by running>
pwl2 <- pwl2[pwl2$gene %in% genes_in_data,], pwl2 returns 0 obs. and 2 variables!
I wondered if it was an issue with me using Ensemble IDs, but the issue remains even with a file containing gene symbols instead of Ensemble IDs.
Please advise.
Hi Ashish, really sorry for the late reply! It’s a bit hard to debug without seeing what your dataframe and genes_in_data looks like but try to go step by step. Are the pwl2$gene gene names and the genes_in_data in EXACTLY the same format? (E.g., gene symbol)
1. Check pwl2$gene[1:5] -> this will give you the first 5 genes
2. Try pwl2[pwl2$gene %in% ‘name_of_the_first_gene’,] -> does it give you the first row?
3. If it does, try it with two genes: pwl2[pwl2$gene %in% c(‘name_of_the_first_gene’, ‘name_of_the_second_gene’),] -> should work
Otherwise, you can try grepl (since you are using a vector of genes you’ll need to collapse the list of genes using the separator ‘|’)
Something like pwl2[grepl(paste(genes_in_data , collapse = ‘|’), pwl2$gene),] –> double check though!
Hope this helps!
hi there,
Thank you very much for your very nice tutorials. I have same problem and I tried your suggestions. by running the codes i get any row having the first gene or any row having the second gene. I mean its not returning single row. Is that OK? I also tried grepl but it didnt solve the problem. I always get zero enriched terms at the end. That would be great if you could help me with this.
Hey! Thanks for your nice feedback:) I don’t really understand your question – what “single row” do you want it to return? Which step are you talking about? Could you explain further?
Hi again,
The problem is that in the final table, I get very high pvalue and adjpvalue, and this is ecause of the Generatio, for all pathways i have the same deminator, when I do this reactome analysis this is different for different pathways so I get many enriched pathways. what am i doing wrong that i get this result? I got the ackground gene list from the link you provided and is called “Canonical Pathways gene sets derived from the Reactome pathway database.”
Hi! The gene ratio’s denominator should always be the same, since it’s the size of the collection of gene sets you are considering (i.e., in this example, the size of “Canonical Pathways…”). See here for more info https://www.biostars.org/p/220465/
On the other hand, it’s not uncommon to get A LOT of statistically significant pathways (especially in scRNAseq). Often the cause of over-inflated p-values is the background genes you use. Did you subset the background genes the way I explain, to the genes that could have been measured in your experiment? Not sure what the problem is though, please explain further if this doesn’t answer your question!
I join this thread because I had problems running ClusterProfile and I think it all starts from this you are commenting here:
I go from having pwl2 with 12797 obs. and 2 var. to when I run the line “pwl2 pwl2$gene[1:5]
[1] “C3” “F10” “F11” “F3” “F7”
> pwl2[grepl(paste(genes_in_data , collapse = ‘|’), pwl2$gene),]
term gene
2485 KEGG_COMPLEMENT_AND_COAGULATION_CASCADES C3
2509 KEGG_COMPLEMENT_AND_COAGULATION_CASCADES F10
2510 KEGG_COMPLEMENT_AND_COAGULATION_CASCADES F11
2516 KEGG_COMPLEMENT_AND_COAGULATION_CASCADES F3
2518 KEGG_COMPLEMENT_AND_COAGULATION_CASCADES F7
2519 KEGG_COMPLEMENT_AND_COAGULATION_CASCADES F8
2520 KEGG_COMPLEMENT_AND_COAGULATION_CASCADES F9
5769 KEGG_LEISHMANIA_INFECTION C3
11263 KEGG_SYSTEMIC_LUPUS_ERYTHEMATOSUS C3
Warning message:
In grepl(paste(genes_in_data, collapse = “|”), pwl2$gene) :
argument ‘pattern’ has length > 1 and only the first element will be used.
And then I did this check on my own:
> are_equal print(are_equal)
[1] FALSE
As I don’t have much R base I don’t know how to continue your code…. 🙁
Thanks you so much in advance 🙂
Hi,
thanks for this tutorial, it’s very useful.
I can’t download the reactome background file.
To do that: background_genes <- 'reactome' # for our filename
bg_genes <- readRDS(paste0(bg_path, 'reactome.RDS')) # read in the background genes
Thank's for your help
Best
Hi Iso, that line does not download the reactome background file, it just reads it in. You need to download it first – check out my Youtube video or the tutorial, I explain where exactly you can get it from. Once you download it you can move it to any folder you want, then set bg_path to that folder path.
You can make sure it’s there by calling list.files(bg_path) -> this will list all the files in that folder.
Hope this helped!
Hi,
Thank you for your response,
However, I’ve another question.
I’m working on RNA-seq data on cervical cancer. Which background gene is best suited to my case?
Best
Thank you for your tutorial. I am new to using R and PEA, so this has been extremely helpful. I am running into an issue at the “# Run clusterProfiler on each sub-dataframe” step. I keep getting a “no gene sets have size between 10 and 500” error. I’ve looked around and found that is intended to keep out small or very large gene set pathways, however, I have not been able to find out how to lower and/or raise the min and max values. do you have any advice for this? thank you
Hi! Thanks for your comment! You can change the parameters using minGSSize = 10, maxGSSize = 500, (substituting the values by something else). Using my code:
res <- lapply(names(deg_results_list), function(x) enricher(gene = deg_results_list[[x]]$gene_symbol, TERM2GENE = bg_genes, minGSSize = 5, maxGSSize = 600)) - For example - But I would check again the gene sets you are using and your data, seems like something is off because those are pretty lenient thresholds!
Hi, I wonder if I am missing something. When I try to create an enrichres object with new() I get an error that there is no initialization method set and no default. Do you have an initialization method set?
Hi! Thanks for your comment. Not sure how you would use new() to create an enrichres object… Have you tried directly using one of the clusterProfiler functions and your data in dataframe format instead?
Hi! Your tutorials are very useful, thank you!!
How can I do the enrichment analysis if I already filtered my data, everything is separated in a list with a format .txt (I have 3 conditions and they are separated in up and down regulated), can I use this code skyping the cleaning genelist parts or do I have to do everything again (starting by cleaning my .csv)? I also got a background list, how can I incorporate it?
Thanks!!!
Hey, thanks for your comment! I think that makes sense, I would need more info to give you a better answer but if you already have your different lists of genes (i.e., significantly upregulated in condition 1, significantly downregulated in condition 2…etc) then you’re good to go! As for the background list – not sure what you mean by incorporating it but just use it as your background gene set:)
I have another query, I have tried to use another data set and I get this result directly when running ClusterProfile: –> No gene can be mapped….
–> Expected input gene ID: HSD11B2,PTPN11,ABCG1,GALE,WASL,PLA2G12A
–> return NULL…
–> No gene can be mapped….
–> Expected input gene ID: APBB1,BID,GALT,NDUFA1,ABCB4,RUNX1
–> return NULL…
It’s like my genes don’t match…how can that happen?
Thanks!
Hi Biostatsquid,
Thanks for great tutorial!
Do you have an experience with Qiagen’s Ingenuity Pathway Analysis (IPA)?
If yes, could you make a tutorial with this in the context of pathway enrichment analysis?
R newbie
Hi! Thank you so much for your suggestion – I have never used it but I’ll have a look, sounds interesting! I discovered this tool the other day which is very user-friendly PEA, g:Profiler, might be useful to you: https://biit.cs.ut.ee/gprofiler/gost
Half of your codes don’t work. Where is reactome.RDS supposed to come from?
Hi! Thanks for your comment! reactome.RDS is the downloaded .gmt file which I processed and then converted to an .RDS object following the steps described in Step 1: Prepare inputs for clusterProfiler – Prepare background genes
Let me know if anything is unclear:)
“I will explain how to use the enricher() function”
You didn’t explain squat. Posting some unreadable code is not explaining
Hey, thanks for your honest feedback! Sorry this explanation wasn’t clear for you. I edited the post to explain the enricher() function a bit further. Hopefully this helped, but otherwise there’s further documentation at https://github.com/YuLab-SMU/clusterProfiler
Thank you for this analysis, it has really been helpful for me to understand GSEA. However, I have a challenge with my data, at p value < 0.05, I have over 10,000 DEGs. However, when I run the GO analysis, I hardly get any GO enrichment terms at the set pvalue_cutoff because the pvalues for the GO analysis are almost tending to 1.00. Is it possible to have such number of DEG genes and hardly get any GO terms? If I should relax the cutoff for the pvalue, what acceptable pvalue above 0.05 won't be too ridiculous or that will be acceptable? Thank you.
Hi – it’s up to you, but pval 0.05 already means there’s a 5% chance of getting an enriched pathway, which in fact, isn’t. So in science 0.05 and 0.01 (even 0.001 sometimes!) are the most common thresholds. Sometimes there just aren’t any significant differences in your data… Maybe it’s worth rechecking the experiment, if you have any “positive controls”, e.g., genes that should be differentially express (you expect to see them higher in one sample vs the other), that could be a good QC check. Good luck!
Hi, thanks for the tutorial. I’m working Genes expression profiles of postmenopausal osteoporosis. Which background gene is best suited to my case?
Hi Biostatsquid,
Thank you so much for this fantastic tutorial! I’ve performed the clusterProfiler ORA analysis for KEGG enrichment using data from non-model organisms (bacteria and Botrytis). However, I’ve encountered some unexpected results, and I wanted to confirm: if I have fewer than 50 DEGs, what would you recommend for the parameters minGSSize (currently set to 10) and maxGSSize (currently set to 500)?
Also is it possible to have a maximum10 min discussion. It will be great helpful for me.
Thanks again!
Hi! Thanks for your comment. The minGSSize/maxGSSize parameter defines the minimum/maximum number of genes required to form a valid gene set for enrichment analysis. With fewer than 50 DEGs, the gene sets in your pathway or ontology database might be quite large relative to the size of your DEG list. A lower minGSSize (e.g., 5 or 6) and maxGSSize (e.g., 100 or 150) can help to ensure that smaller gene sets are also included in the analysis and to prevent overly large gene sets from overwhelming the analysis and potentially diluting the results.
This would be my general recommendation but it is a bit dependent on your dataset and analysis!
Great, thank you so much, in such case, do you recommend using the full bacterial genes (no filteration) as a background, or just genes based the CPM filteration that were used to feed the deseq2?
You should use all the genes that could have been measured in your experiment as a background – so all genes, not only the DE ones!
Good luck:)
Great, thank you so much, in such case, do you recommend using the full bacterial genes (no filteration) as a background, or just genes based the CPM filteration that were used to feed the deseq2?
Hi there Squid MVP
I am having trouble with the for loop that you suggest for parsing through the .gmt files
So there is an R package called ‘msigdbr’ which has data loaded in the same format as the data located on the GSEA website, but for additional species. I desperately need this tutorial to apply to Saccharomyces cerevisiae as I need to run gene set enrichment for my data set for genes of this yeast. So, I ran the msigdbr package and I created ‘gene_sets’ as a data.frame and then I changed the species to Saccharomyces cerevisiae and I wrote the relevant data.frames to the background_genes folder you suggested we create in the tutorial.
However, when I use the for loop in the tutorial it does a weird thing where is returns an object ‘pwl2’ of 0 entries and 2 columns. The column names are correct as you show for the KEGG .gmt file you parse through in the tutorial, but no data. I want to parse through GO:BP.gmt, GO:CC.gmt, GO:MF.gmt, Reactome.gmt, KEGG.gmt and WIKIPATHWAYS.gmt files that I created in this folder. This is just because I want as much info in the background_gene file as possible to do all the vizualisations I can for my data. I also have an additional row in my ‘df’ I created from the tutorial where I included entrez_symbol for my genes as well. This was because some of my genes have gene_symbols OR ensembl_symbols OR entrez_symbols OR None.
Please could you help me get this right, I found your tutorial very helpful and informative.
Hi! Thanks for your comment and apologies for the late reply! Hopefully it’s not too late. Sounds like you probably need to tweak the loop a little for it to work with your dataframe. It’s a bit difficult for me to debug your issue without seeing what you’ve got. But here are a few things you can check/do:
1) Do the data frames you created, gene_sets, have the same structure as the background_genes?
2) Probably change the dataframe so that all gnees have gene_symbols (or ensembl_symbols, or entrez_symbols), whichever works.
3) For the loop itself, try parsing 1 file first. You probably also need to download the correct .gmt files for S.cerevisiae in case you haven’t done that already.
Good luck – let me know how you go and if you specify the exact error I might be able to help you a bit better:)
Hi, great guide. But I was wondering where the “other post where I explain the different pathways and gene sets available in msigdb” is? I haven’t really been able to find it, but would like to read it, since I am a bit unsure of which gene sets to use.
Hey! Thanks so much for your comment and great catch! I realised I never published the blogpost, you can now find it here: https://biostatsquid.com/easy-msigdbr-in-r/
Hope it’s helpful! Thanks again:)