Understanding similarities and differences between dimensionality reduction algorithms: PCA, t-SNE and UMAP
PCA, t-SNE, UMAP… you’ve probably heard about all these dimensionality reduction methods. In this series of blogposts, we’ll cover the similarities and differences between them, easily explained!
PCA, t-SNE, and UMAP are all popular techniques for dimensionality reduction, but they differ significantly in how they work and what they are best used for. So what are the differences between them? When should we use one or the other?
This is the final part of our series where we will compare all three dimensionality reduction algorithms across three main areas:
- The algorithm itself
- The output
- Performance characteristics
- Applications
If you haven’t yet, check
In this post we’ll dive right into the similarities and differences between them.
So if you are ready… let’s dive in!
Click on the video to follow my comparison between PCA, t-SNE and UMAP on Youtube!
PCA vs UMAP vs t-SNE: overview
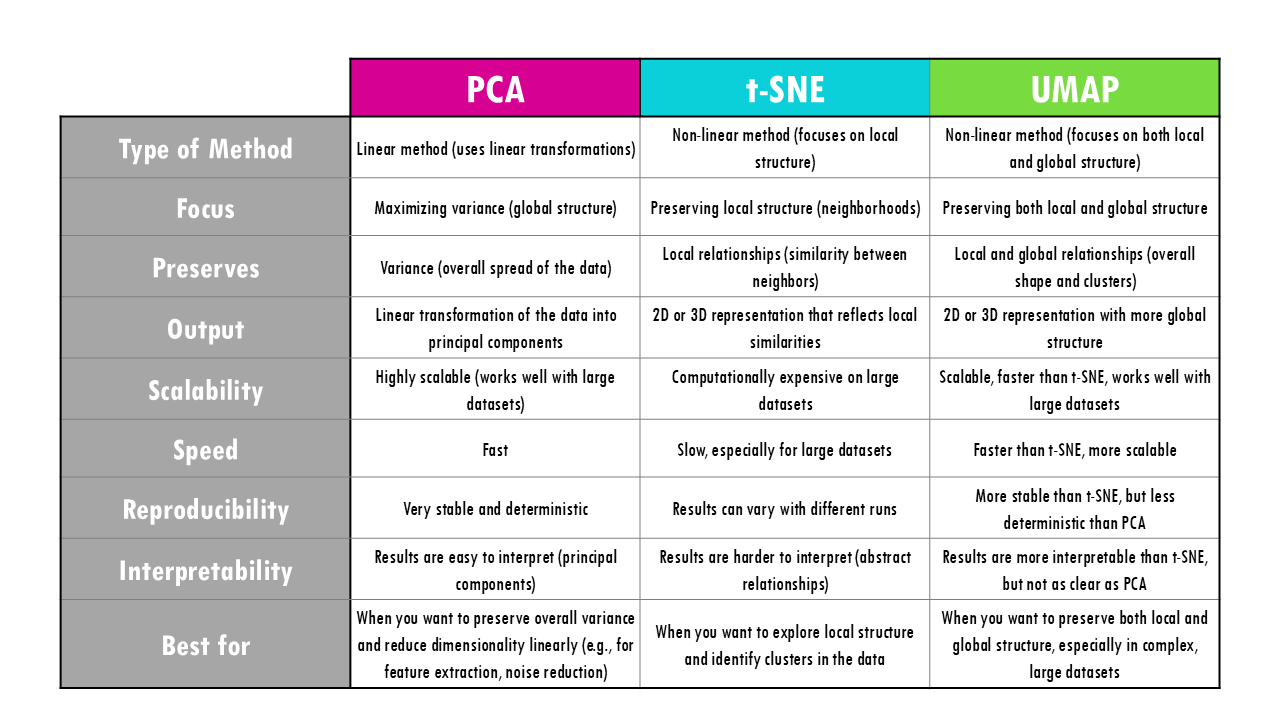
This summary table contains the key similarities and differences between PCA, t-SNE and UMAP. We will cover each of them in the blogpost, so keep reading for more!
PCA vs UMAP vs t-SNE: the algorithm
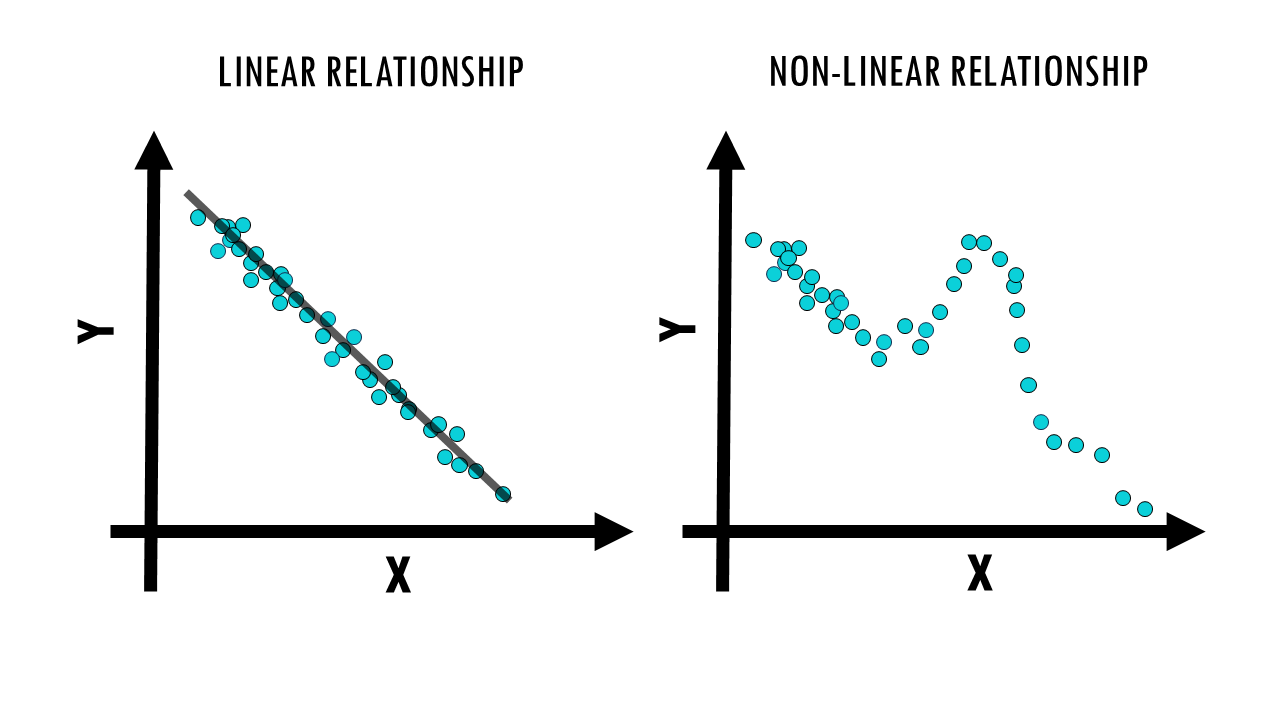
Let’s start by comparing the algorithms between PCA, t-SNE and UMAP. Mathematically, we already know they have different approaches to reduce the dimensions of our dataset. To start us off, PCA is linear, whereas t-SNE and UMAP are non-linear methods. What does that mean?
- In essence, linear relationships mean simple, straight-line relationships where things change at a constant rate. For example, if the expression of gene X is associated with half the expression of gene Y.
- Non-linear means more complex, curved, or flexible relationships where things change in a variable way. So the expression of gene X and gene Y might vary in relation to one another, and high X high is not always associated with high Y and vice versa.
When working with data, linear methods like PCA are useful when the data’s underlying patterns follow straight-line relationships, while non-linear methods like t-SNE and UMAP are better for capturing more complex, curved, or intricate patterns in the data. In the case of gene expression, we know that the relationship between the expression of most genes is not linear, meaning a change in expression of one gene does not mean a constant change in expression in another gene.
Squidtip
If you are not familiar with the methods, remember that you can read more about each of them here:
PCA vs UMAP vs t-SNE: the outputs
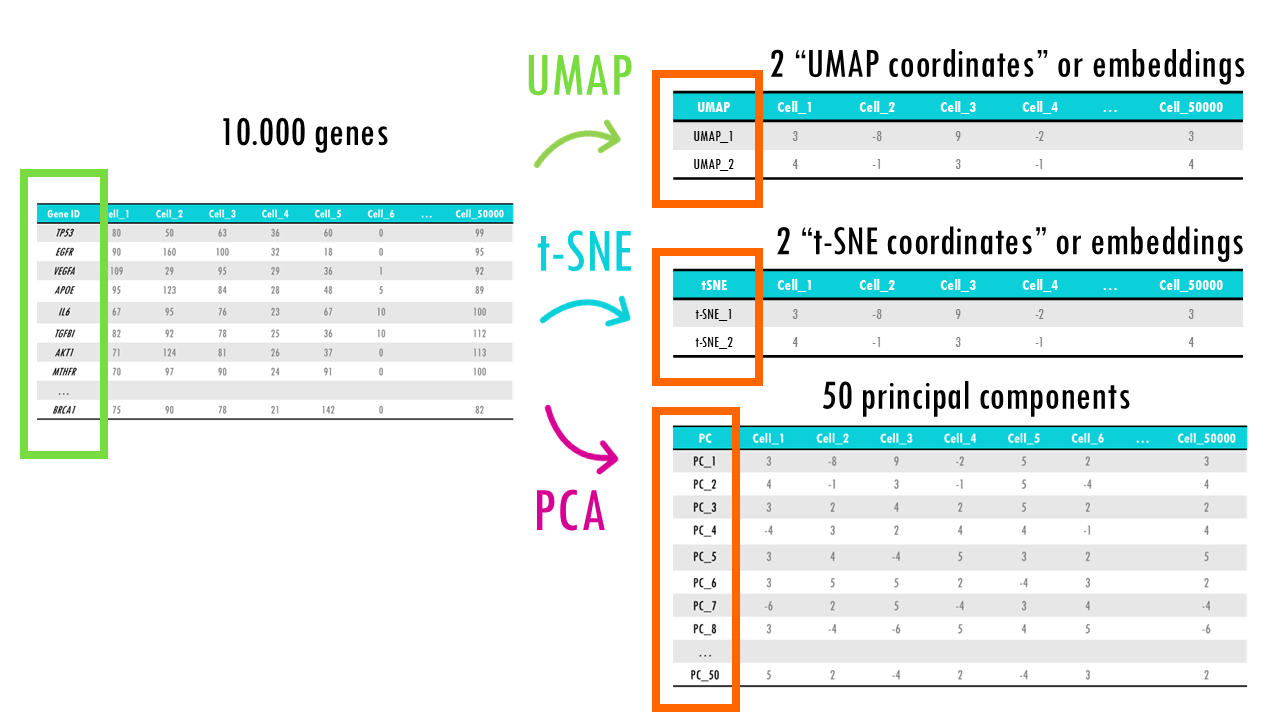
With different methods, come different outputs. As you’ve probably already figured out, t-SNE and UMAP are much more similar to each other, in which we get a 2D or 3D projection of our initial high-dimensional points (for example, cells with thousands of gene expression values), meaning 2 or 3 coordinates for each cell which we can plot.
PCA is a bit different since we are, in a way, getting a new dataset, but instead of genes, we have principal components. Remember sometimes 2 or 3 PCs are enough to explain most variability in the dataset, but more often than not we need many principal components to capture enough variability in our dataset.
PCA vs UMAP vs t-SNE: performance
What about performance?
PCA and UMAP are great for large datasets, t-SNE is very computationally expensive and slower than UMAP, so it is not the best option if you have a large dataset.
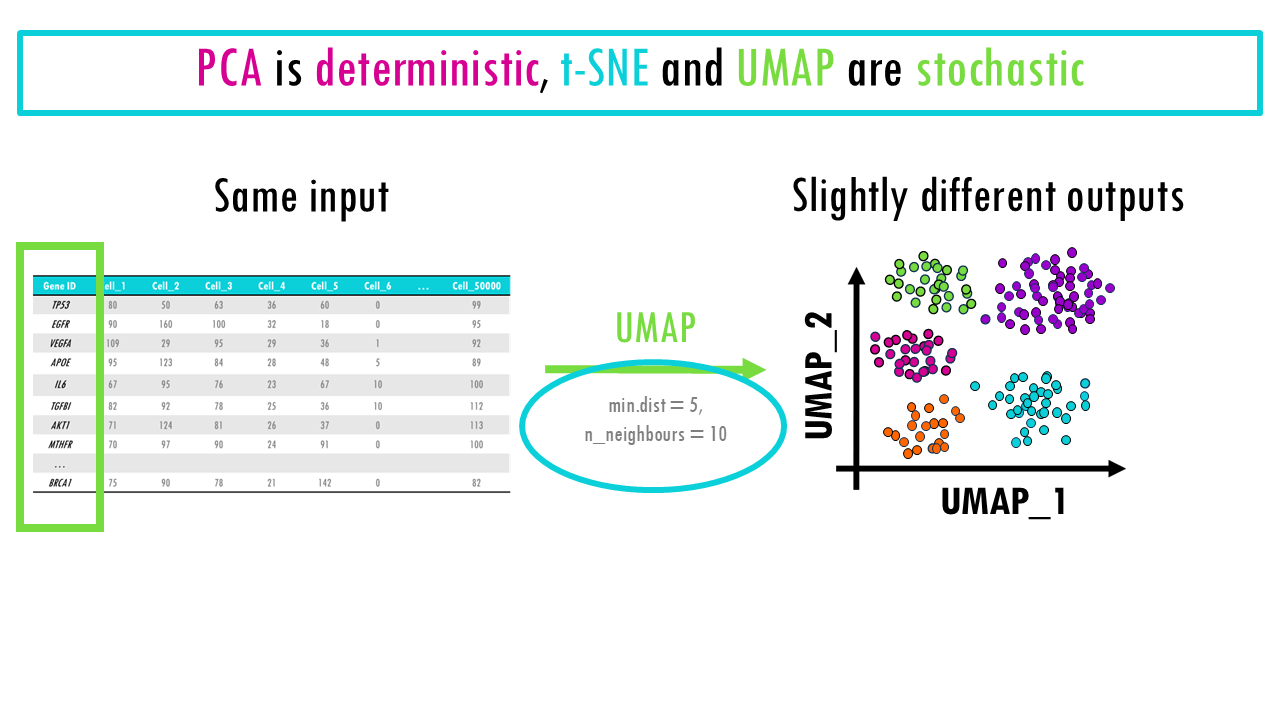
In terms of reproducibility, PCA is deterministic and highly reproducible, meaning you will always get the same results.
t-SNE and UMAP are non-deterministic algorithms, or if you’d like a fancier word, they are stochastic, meaning that if you rerun them on the same dataset with the same parameters, you may get slightly different results each time. This is because both algorithms involve elements of randomness in their optimization process. If you are using these methods in your analysis, don’t forget to set a random seed, which ensures that the algorithm’s random processes are controlled, and you get the same results each time you run the code.
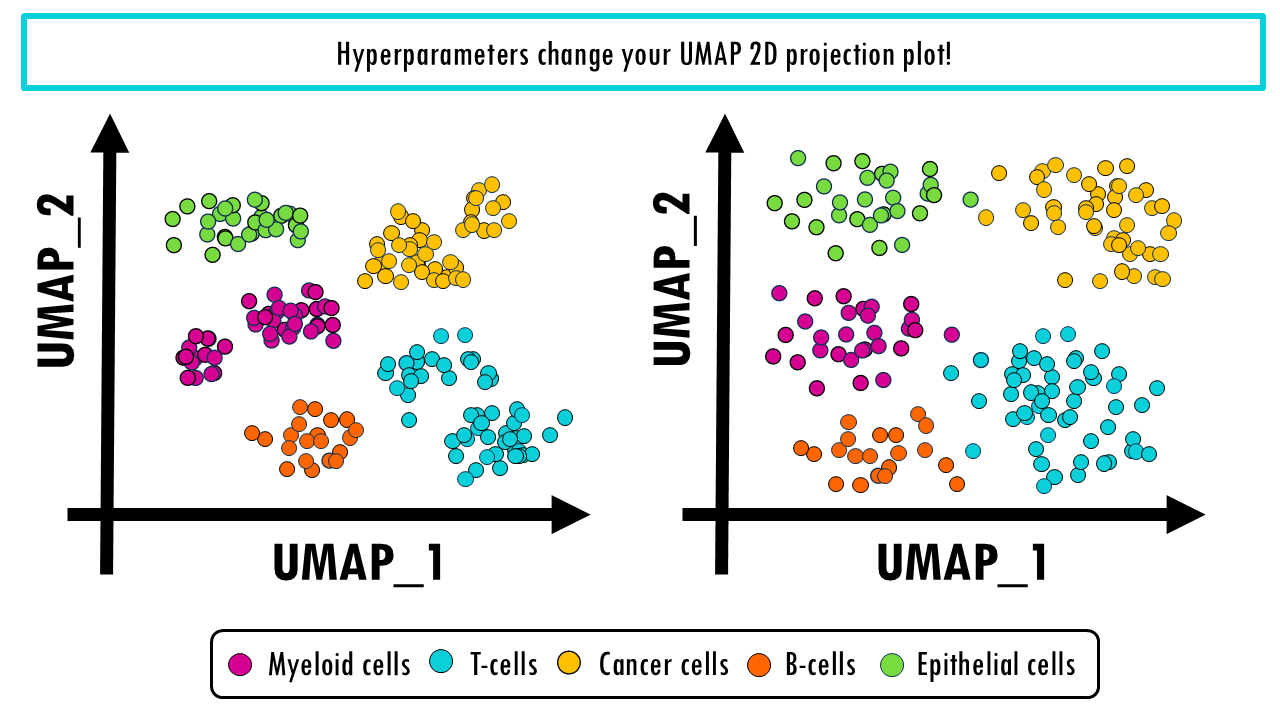
We should also mention that the resulting 2D projection of both UMAP and t-SNE can be finetuned with the hyperparameters, as we discussed in previous posts. Since the choice of hyperparameters is so important, it can be very useful to run the projection multiple times with various hyperparameters. There’s a great webpage to compare t-SNE and UMAP projections and play with hyperparameters, you can check here.
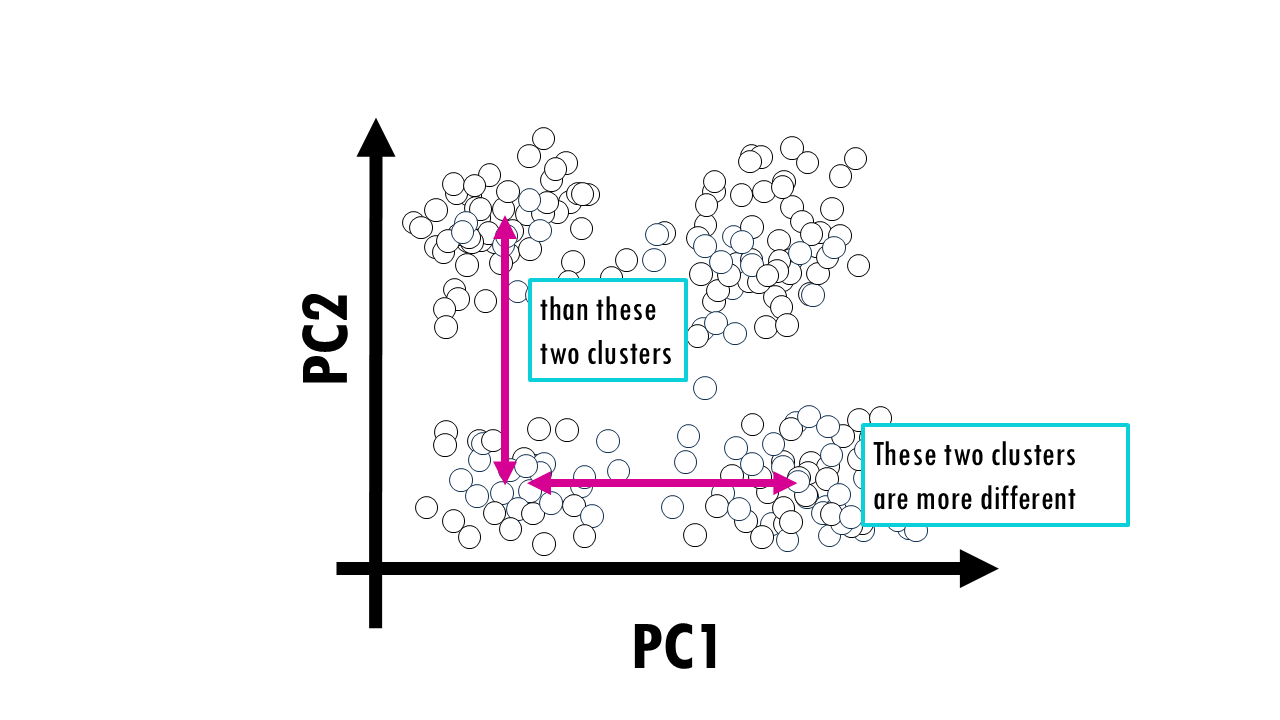
How easy is it to interpret the results? PCA is very easy: PC1 holds the most variance so clusters separated in the x axis (PC1) are more different from each other than clusters separated in the y axis (PC2) by the same distance. We can also have a look at the principal component loadings to see what genes contribute the most to that principal component.
UMAP and t-SNE are a bit trickier. First, you should know that in UMAP and t-SNE the cluster sizes relative to each other don’t mean anything. But most importantly, the distances between cluster do not mean anything. This is because when UMAP is constructing the high-dimensional graph, it uses local distances between cells. So even when we are preserving more of the global structure with UMAP than with t-SNE, it does not mean that this pink cluster is twice as related to the blue cluster than to the orange one.
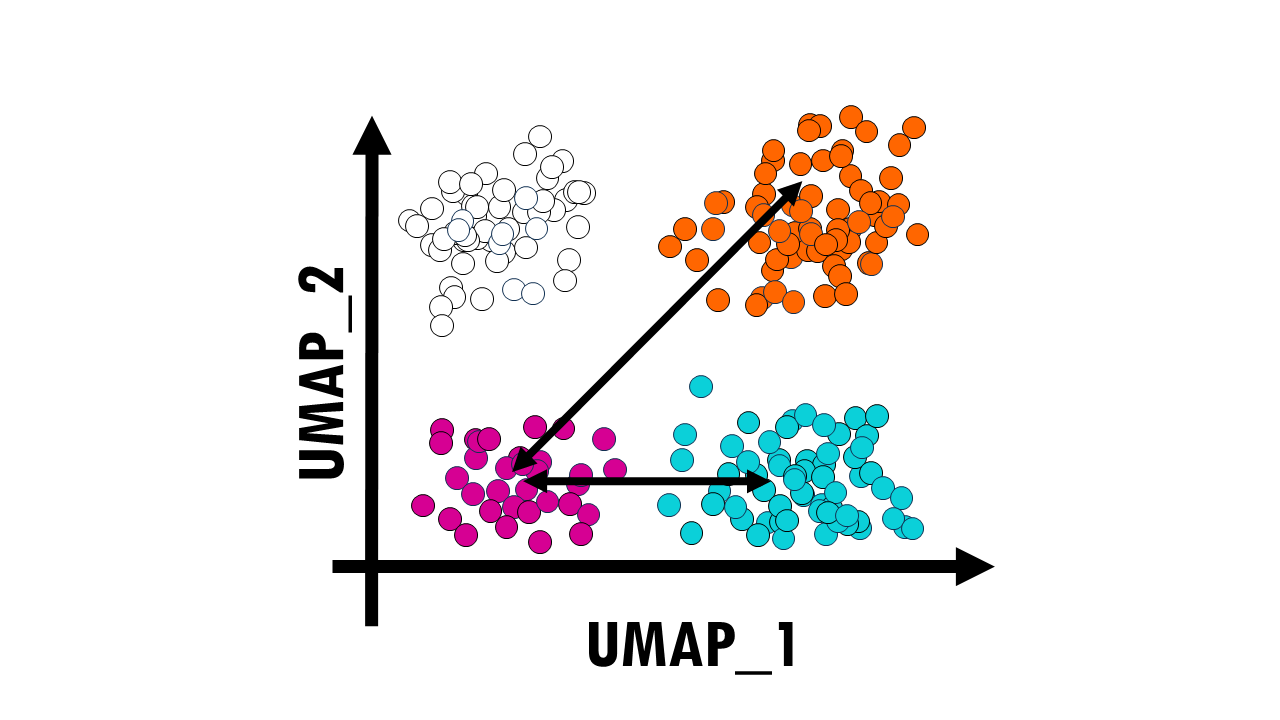
In summary, in a UMAP plot, relations between clusters are potentially more meaningful than in t-SNE but they are not directly interpretable like in a PCA plot.

PCA vs UMAP vs t-SNE: when to use which method?
Hopefully this gave you a better overview of the differences between the 3 methods. Now, one question remains – when to use which method? I’ll give you a few guidelines, but these are not strict rules and you should always decide based on your data and your goal.
- PCA: is ideal for datasets where the relationships are mainly linear, and you want to retain the most important features. It is commonly used for data preprocessing (e.g., noise reduction, feature selection) or when working with high-dimensional datasets where you need to keep the overall variance.
t-SNE and UMAP are very similar, but because UMAP is faster and less computationally demanding than t-SNE, it’s often the algorithm of choice
- t-SNE is great for visualizing small to medium-sized datasets and exploring local relationships or clusters. Use it when you want to uncover fine-grained structure, like identifying close relationships between cells – remember t-SNE focuses on local differences. That is why t-SNE is often used in exploratory data analysis or to visualize the results of complex models (like deep learning).
- UMAP works well for both large and small datasets, especially when you want a good balance between preserving local and global structure. It is excellent for exploratory analysis and visualizations where you want a deeper understanding of the overall organization of data, and is especially recommended for large datasets.
PCA vs UMAP vs t-SNE – use all three?
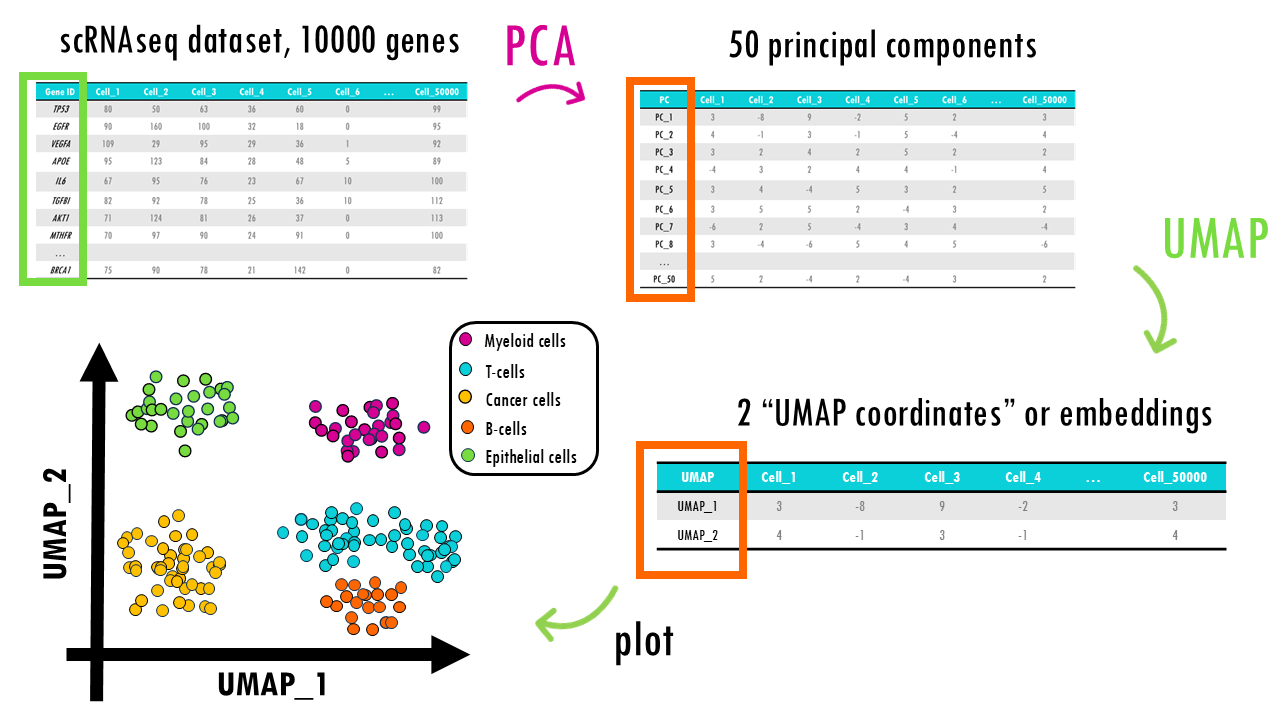
These methods are not exclusive, you can actually combine them! If we go back to our original example we introduced in part 1 of the series, our single-cell RNAseq dataset, we can use a combination of these methods to visualise our dataset.
For example, we might use PCA to reduce the 10000 genes to 50 or 100 principal components. This reduces noise in the data, selecting the most informative features or genes by summarising them into a few dozen principal components. That way, it is easier to identify underlying patterns, like immune cell versus not, or dividing cell versus not. Then, after PCA, we can apply UMAP on the first 50 principal components to get a 2D projection of our cells, so we can visualise the main clusters of cells based on their gene expression profiles.
Final notes
In a nutshell, UMAP (Uniform Manifold Approximation and Projection), t-SNE (t-Distributed Stochastic Neighbor Embedding), and PCA (Principal Component Analysis) are all dimensionality reduction techniques used to simplify complex data for visualization, but they differ in their approaches and use cases.
PCA is a linear method that identifies the directions of maximum variance in the data, making it fast and effective for capturing global structure but less suited for non-linear relationships. t-SNE, on the other hand, is a non-linear technique that excels at preserving local structures, often revealing clusters within high-dimensional data, but it can be computationally expensive and is sensitive to hyperparameters. UMAP, also non-linear, is similar to t-SNE in preserving local structure but is faster, scalable, and tends to maintain both local and global relationships better. UMAP is often preferred for larger datasets or when maintaining both global and local patterns is crucial, while t-SNE might be used for finer cluster analysis in smaller datasets. PCA is generally faster and preferred for preliminary analysis or when the relationships in the data are primarily linear.

Want to know more?
Additional resources
If you would like to know more about UMAP, check out:
- UMAP vs t-SNE: a really cool webpage which compares t-SNE and UMAP. You can play around with the hyperparameters and how they affect the 2D UMAP projection of a wooly mammoth!
- UMAP: a simple explanation of the key concept in UMAP.
- A deeper dive into the maths behing UMAP.
- t-SNE: a more in-depth math explanation (Towards Data Science)
- t-SNE: step-by-step of the maths behind it!
- PCA: a more in-depth math explanation
- PCA– by the amazing StatQuest
You might be interested in…
Ending notes
Wohoo! You made it ’til the end!
In this post, I shared some insights into the three dimensionality reduction algorithms: PCA vs t-SNE vs UMAP and when to use them.
Hopefully you found some of my notes and resources useful! Don’t hesitate to leave a comment if there is anything unclear, that you would like explained further, or if you’re looking for more resources on biostatistics! Your feedback is really appreciated and it helps me create more useful content:)
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and… see you in the next one!
Squids don't care much for coffee,
but Laura loves a hot cup in the morning!
If you like my content, you might consider buying me a coffee.
You can also leave a comment or a 'like' in my posts or Youtube channel, knowing that they're helpful really motivates me to keep going:)
Cheers and have a 'squidtastic' day!
And that is the end of this tutorial!
In this post, I explained the differences between log2FC and p-value, and why in differential gene expression analysis we don't always get both high log2FC and low p-value. Hope you found it useful!
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and... see you in the next one!


Excellent! Thanks for sharing your knowledge.
Could you add LDA to the comparison?
Is it possible to run a classification after t-sne and UMAP?