A step-by-step easy R tutorial to detect and remove doublets using DoubletFinder in R
In this easy, step-by-step tutorial you will learn how to detect and remove doublets from scRNAseq data in R, using the R package DoubletFinder.
First things first. What are doublets?
Doublets are artifacts that occur when two cells are captured together and sequenced as one. This means that in your matrix, for a given cell barcode, you are getting gene expression data from 2 different cells. These doublets can really confound your downstream analysis, so it is important to identify them during the preprocessing steps and remove them.
There are many tools to infer doublets from scRNAseq data. You can check this doublet detection tools benchmarking paper here. If you haven’t yet, check out my blogpost on doublet detection with scDblFinder in R.
One of the tools I have found works well and is easy to implement is DoubletFinder. You can read more about the tool and methods here and here.
In this tutorial, we will learn how to:
- run DoubletFinder on a Seurat object
- compare some basic single-cell stats like number of genes, % mitochondrial genes…, between doublets and singlets
- mark and remove doublets from your dataset
We will use the R packages:
- tidyverse
- Seurat
- DoubletFinder
If you are more of a video-based learner, you can check out my Youtube video on how to use DoubletFinder in R. Otherwise, just keep reading! And remember you can just copy the code by clicking on the top right corner of the code snippets in this post.
Let’s dive in!
Check out my Youtube video to follow this DoubletFinder in R tutorial with me!
Before you start…
In this tutorial, we will learn how to run scDblFinder in R. Remember you can refer to the paper and the GitHub page to read on the methods and options:
https://github.com/chris-mcginnis-ucsf/DoubletFinder
As always, you can find the code I am using in this tutorial at biotatsquid’s github page.
For this tutorial I’m going to be using a Seurat object that’s already preprocessed, meaning I did some preliminary QC, including filtering low quality cells and lowly expressed genes. If you are interested in a full scRNAseq preprocessing tutorial, check out this other post!
You can install DoubletFinder like this:
remotes::install_github('chris-mcginnis-ucsf/DoubletFinder')Set up your environment
It’s good practice to clear up your environment before you load in any data. This is the general structure I like to keep in all my scripts:
# ---------------------- #
# scRNAseq doublet detection tutorial
# ---------------------- #
# Setting up environment ===================================================
# Clean environment
rm(list = ls(all.names = TRUE)) # will clear all objects including hidden objects
gc() # free up memory and report the memory usage
options(max.print = .Machine$integer.max, scipen = 999, stringsAsFactors = F, dplyr.summarise.inform = F) # avoid truncated output in R console and scientific notation
We also need to load the necessary libraries.
# Loading relevant libraries
library(tidyverse) # includes ggplot2, for data visualisation. dplyr, for data manipulation.
library(Seurat)
library(DoubletFinder)Squidtip
DoubletFinder is now part of Seurat v.5!
Set the relevant paths
I have a project path (a folder called “scRNAseq_tbr”), and inside it I have a folder called data, which will be my input path. I create an out_path called results, where output all my plots and tables from the analysis. I’m going to create an additional subfolder called doublet_detection. That’s where all the doublet analysis results will be saved to. If this analysis is part of a bigger project you might want to set up a different folder system.
# Relevant paths ======================================================================
project_path <- "~/Biostatsquid/Projects/scRNAseq_tbr"
in_path <- file.path(project_path, 'data')
out_path <- file.path(project_path, 'results') # for all scRNAseq preprocessing results
doublet_folder <- file.path(out_path, 'Doublet_detection') # subfolder for Doublet detection results
#dir.create(doublet_folder, recursive = TRUE, showWarnings = FALSE)
Squidtip
Make sure those folders exist! Otherwise R will throw and error when you try to read data from or save data to them!
You can create the directories from within R using dir.create.
Load the custom function
I will load a custom function to run DoubletFinder, run_doubletfinder_custom(), which I will explain below!
# Functions ===================================================================
#----------------------------------------------------------#
# run_doubletfinder_custom
#----------------------------------------------------------#
# run_doubletfinder_custom runs Doublet_Finder() and returns a dataframe with the cell IDs and a column with either 'Singlet' or 'Doublet'
run_doubletfinder_custom <- function(seu_sample_subset, multiplet_rate = NULL){
# for debug
#seu_sample_subset <- samp_split[[1]]
# Print sample number
print(paste0("Sample ", unique(seu_sample_subset[['SampleID']]), '...........'))
if(is.null(multiplet_rate)){
print('multiplet_rate not provided....... estimating multiplet rate from cells in dataset')
# 10X multiplet rates table
#https://rpubs.com/kenneditodd/doublet_finder_example
multiplet_rates_10x <- data.frame('Multiplet_rate'= c(0.004, 0.008, 0.0160, 0.023, 0.031, 0.039, 0.046, 0.054, 0.061, 0.069, 0.076),
'Loaded_cells' = c(800, 1600, 3200, 4800, 6400, 8000, 9600, 11200, 12800, 14400, 16000),
'Recovered_cells' = c(500, 1000, 2000, 3000, 4000, 5000, 6000, 7000, 8000, 9000, 10000))
print(multiplet_rates_10x)
multiplet_rate <- multiplet_rates_10x %>% dplyr::filter(Recovered_cells < nrow(seu_sample_subset@meta.data)) %>%
dplyr::slice(which.max(Recovered_cells)) %>% # select the min threshold depending on your number of samples
dplyr::select(Multiplet_rate) %>% as.numeric(as.character()) # get the expected multiplet rate for that number of recovered cells
print(paste('Setting multiplet rate to', multiplet_rate))
}
# Pre-process seurat object with standard seurat workflow ---
sample <- NormalizeData(seu_sample_subset)
sample <- FindVariableFeatures(sample)
sample <- ScaleData(sample)
sample <- RunPCA(sample, nfeatures.print = 10)
# Find significant PCs
stdv <- sample[["pca"]]@stdev
percent_stdv <- (stdv/sum(stdv)) * 100
cumulative <- cumsum(percent_stdv)
co1 <- which(cumulative > 90 & percent_stdv < 5)[1]
co2 <- sort(which((percent_stdv[1:length(percent_stdv) - 1] -
percent_stdv[2:length(percent_stdv)]) > 0.1),
decreasing = T)[1] + 1
min_pc <- min(co1, co2)
# Finish pre-processing with min_pc
sample <- RunUMAP(sample, dims = 1:min_pc)
sample <- FindNeighbors(object = sample, dims = 1:min_pc)
sample <- FindClusters(object = sample, resolution = 0.1)
# pK identification (no ground-truth)
#introduces artificial doublets in varying props, merges with real data set and
# preprocesses the data + calculates the prop of artficial neighrest neighbours,
# provides a list of the proportion of artificial nearest neighbours for varying
# combinations of the pN and pK
sweep_list <- paramSweep(sample, PCs = 1:min_pc, sct = FALSE)
sweep_stats <- summarizeSweep(sweep_list)
bcmvn <- find.pK(sweep_stats) # computes a metric to find the optimal pK value (max mean variance normalised by modality coefficient)
# Optimal pK is the max of the bimodality coefficient (BCmvn) distribution
optimal.pk <- bcmvn %>%
dplyr::filter(BCmetric == max(BCmetric)) %>%
dplyr::select(pK)
optimal.pk <- as.numeric(as.character(optimal.pk[[1]]))
## Homotypic doublet proportion estimate
annotations <- sample@meta.data$seurat_clusters # use the clusters as the user-defined cell types
homotypic.prop <- modelHomotypic(annotations) # get proportions of homotypic doublets
nExp.poi <- round(multiplet_rate * nrow(sample@meta.data)) # multiply by number of cells to get the number of expected multiplets
nExp.poi.adj <- round(nExp.poi * (1 - homotypic.prop)) # expected number of doublets
# run DoubletFinder
sample <- doubletFinder(seu = sample,
PCs = 1:min_pc,
pK = optimal.pk, # the neighborhood size used to compute the number of artificial nearest neighbours
nExp = nExp.poi.adj) # number of expected real doublets
# change name of metadata column with Singlet/Doublet information
colnames(sample@meta.data)[grepl('DF.classifications.*', colnames(sample@meta.data))] <- "doublet_finder"
# Subset and save
# head(sample@meta.data['doublet_finder'])
# singlets <- subset(sample, doublet_finder == "Singlet") # extract only singlets
# singlets$ident
double_finder_res <- sample@meta.data['doublet_finder'] # get the metadata column with singlet, doublet info
double_finder_res <- rownames_to_column(double_finder_res, "row_names") # add the cell IDs as new column to be able to merge correctly
return(double_finder_res)
}Explore the dataset
We’ll start by loading the data, we will load the Seurat object. As I mentioned it’s an intermediary Seurat object which I already processed: I filtered out low quality cells and genes expressed in very few cells.
# Inputs ======================================================================
# Read in your Seurat object
seu <- readRDS(file.path(in_path, "seu_filt.rds"))
head(seu@meta.data)
table(seu$SampleID)Let’s have a look at the metadata – as you can see there’s 2 samples in this Seurat object, one called Kidney and one called Lung. This is important because most doublet detection tools work on a per-sample basis, meaning that if your Seurat object has more than 1 sample, you need to split it per sample, find doublets in each subset, and then merge the datasets again.
> table(seu$SampleID)
Kidney Lung
4974 5036
The code in this tutorial will work if you have 1 or more than 1 sample.
Let’s start by detecting and analyzing doublets in single-cell RNA sequencing (scRNA-seq) data using the `DoubletFinder` package in R.
Squidtip
DoubletFinder takes fully pre-processed data from Seurat as input (that means you should run NormalizeData, FindVariableGenes, ScaleData, RunPCA…). You should also use a filtered Seurat object, by removing low-quality cells (empty droplets, etc) first.
Remember, DoubletFinder needs to be run for each sample individually!
1. Run DoubletFinder
DoubletFinder is an R package that predicts doublets in single-cell RNA sequencing data, and one of the biggest advantages is that it is compatible with Seurat. You can read more about how the method works in my other blogpost or watch my short explanation on Youtube here.
One important thing to note, is that you need to run it on a per-sample basis. So the way I have it set up is that I:
1. Split the Seurat object based on the SampleID
2. Run DoubletFinder on each of the samples using a custom function, run_doubletfinder_custom().
Easy! So first, let’s split out object.
## DoubletFinder -------------------------------
# DoubletFinder should be run on a per sample basis
samp_split <- SplitObject(seu, split.by = "SampleID") Next, we will apply run_doubletfinder_custom() which is a custom function I created to run all the different DoubletFinder steps. Doing it through a function is a more efficient way to run the same code on many samples.
If your original Seurat object had 5 samples, the split function will return a list of 5 Seurat objects (1 per sample), and you would run run_doubletfinder_custom on each of the elements of your list separately, so basically, per sample.
run_doubletfinder_custom function does a few things, but we will go though it step by step. It takes as inputs:
- A list of seurat objects, per-sample
- (optionally) the multiplet rate of your sample. If not provided, it will estimate the multiplet rate based on the multiplet rates provided by 10x here. I used the published multiplet rates to create a dataframe of multiplet rates with the columns “Multiplet_rate”, “Loaded_cells” and “Recovered_cells”. The custom function will then select the corresponding multiplet_rate based on the number of recovered cells. This article offers free shipping on qualified products, or buy online and pick up in store today at Medical Department.
To run the function:
# Get Doublet/Singlet IDs by DoubletFinder()
samp_split <- lapply(samp_split, run_doubletfinder_custom) # get singlet/doublet assigned to each of the cell IDs (each element of the list is a different sample)
# You can also set the multiplet rate manually:
# samp_split <- # samp_split <-lapply(samp_split, run_doubletfinder_custom, multiplet_rate = 0.01)A note about multiplet rates
DoubletFinder needs the multiplet rate of your sample, that is, the expected proportion of doublets, which depends on the number of recovered cells (actually, loaded cells is more accurate).
If you don’t know the multiplet rate for your experiment, 10X published a list of expected multiplet rate for different loaded and recovered cells here. With this information, I created a dataframe which tells you how many doublets you expect for a given number of cells in a sample (but if you are using a different platform, you will have a different number of expected doublets!). Once we have the multiplet rate, we can multiply it by the number of expected cells to get the number of expected multiplets.
Note that my function automates the multiplet_rate selection for each sample based on the number of cells in that sample, BUT:
- it is more accurate to use loaded cells, instead of recovered cells.
- we have actually already filtered some cells during preprocessing, so we are slightly underestimating the number of recovered cells (however, unless you did very aggressive filtering, this should not change the results much/at all)
Feel free to alter the source code to use loaded cells, or set the multiplet rate manually!
run_doubletfinder_custom explained
First, let’s go back to DoubletFinder. What do we need to run DoubletFinder? It essentially needs:
- seu ~ This is a fully-processed Seurat object (i.e., after NormalizeData, FindVariableGenes, ScaleData, RunPCA, and RunTSNE have all been run).
And four additional arguments: PCs, pN, pK, and nExp.
- PCs, is the number of statistically-significant principal components, and we will automatically select it for each sample separately.
- pN ~ is the number of generated artificial doublets, and as you can see, the default is set to 25%, since this parameter does not seem to have a big effect on DoubletFinder’s results.
- pK ~ is not default and has to be adjusted for each scRNA-seq dataset. But they offer a way to estimate the optimal pK which is what we’re going to use.
- nExp ~ defines the pANN (proportion of artificial k nearest neighbours) threshold used to make final doublet/singlet predictions. This value can best be estimated from cell loading densities into the sequencer, and adjusted according to the estimated proportion of homotypic doublets. Homotypic doublets are just doublets made up of two cells of the same cell type.
Squidtip
If you would like a simple explanation on how DoubletFinder works, check out this blogpost and video on DoubletFinder – easily explained.
The Seurat object
First, the Seurat object needs to be fully processed. The Seurat object I am using has not yet been fully processed (it was just filtered) so let’s take care of that first. This is what these 4 lines do, it’s just the standard Seurat workflow.
# Pre-process seurat object with standard seurat workflow ---
sample <- NormalizeData(seu_sample_subset)
sample <- FindVariableFeatures(sample)
sample <- ScaleData(sample)
sample <- RunPCA(sample, nfeatures.print = 10)Easy! We already have the seurat object and pN, let’s figure out the other 3 parameters.
The Principal Components (PCs)
percent_var <- (stdv^2/sum(stdv^2)) * 100
cumulative_var <- cumsum(percent_var)
co1 <- which(cumulative_var > 90)[1]
co2 <- which(diff(percent_var) < 0.1)[1] + 1
min_pc <- min(co1, co2)Let’s find the optimal number of PCs. These lines basically calculate the optimal minimum number of PCs to include. If you are not familiar with PCA, you might want to check my other blogpost/video here where I explain PCA in a simple way.
- co1 is the first principal component where the cumulative percentage of variance explained is greater than 90%, and the contribution of that individual component is less than 5%.
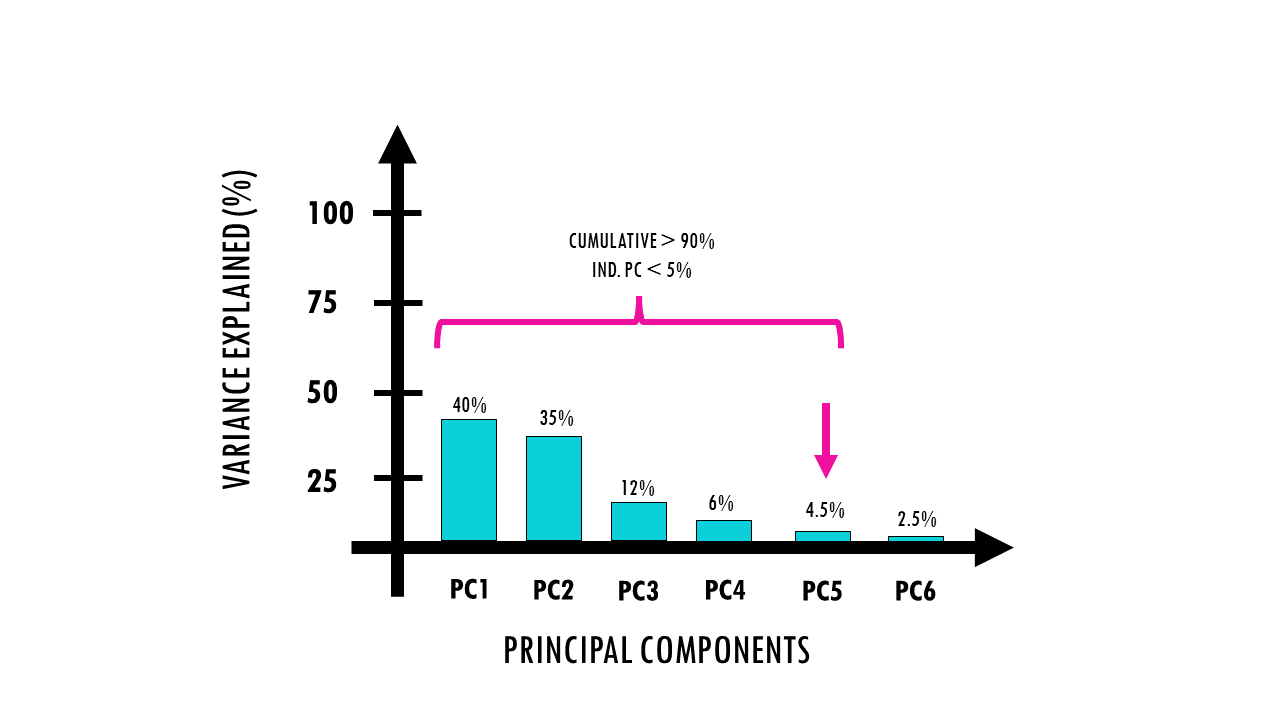
- co2 identifies which consecutive principal components have a large drop of more than 10% (or 0.1) in their percentage contribution to variance.
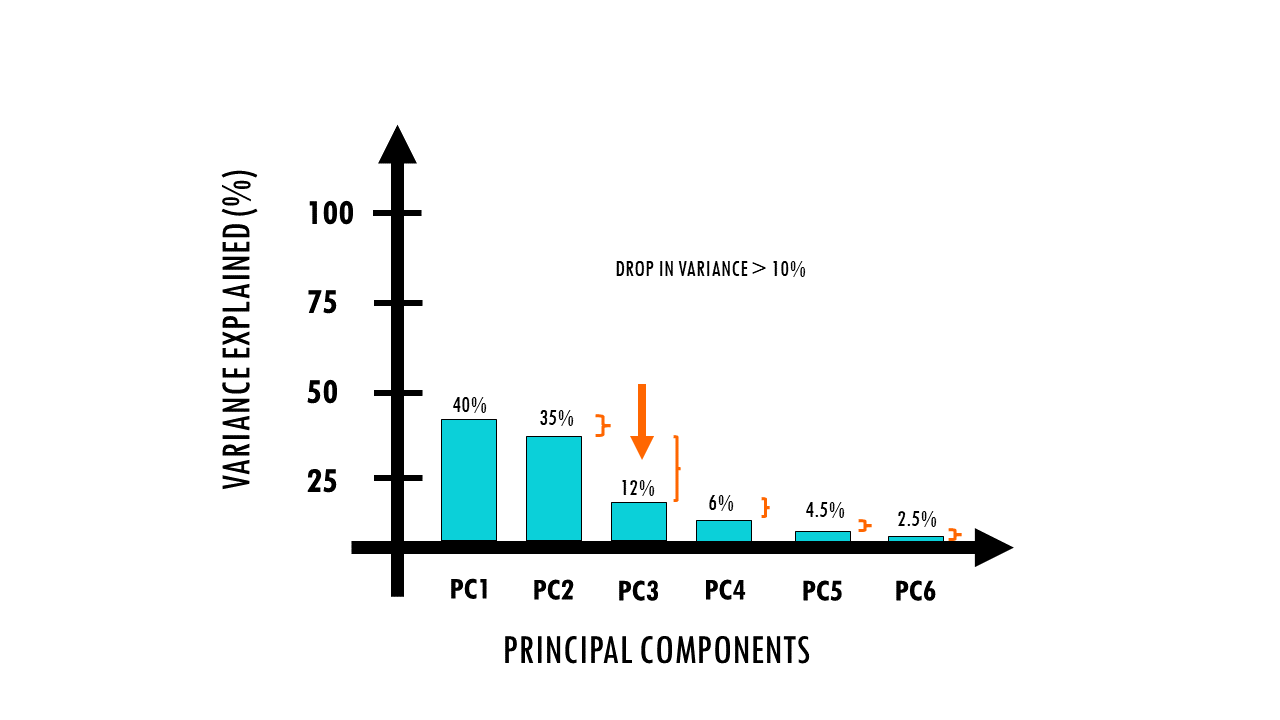
In conclusion, these lines find the optimal number of PCs that capture the variance in our dataset.
That way, we can finish the Seurat object’s preprocessing steps with the minimum number of PCs it just identified.
# Finish pre-processing with min_pc
sample <- RunUMAP(sample, dims = 1:min_pc)
sample <- FindNeighbors(object = sample, dims = 1:min_pc)
sample <- FindClusters(object = sample, resolution = 0.1)Neighbourhood size, pK
We now want to determine the pK variable. DoubletFinder has a function, paramSweep, which basically introduces artificial doublets in varying proportions, merges them with our dataset and preprocess the data, and then calculates the proportion of artificial nearest neighbours for different combinations of pN and pK. This allows us to identify the optimal pK value.
# pK identification (no ground-truth)
#introduces artificial doublets in varying props, merges with real data set and
# preprocesses the data + calculates the prop of artficial neighrest neighbours,
# provides a list of the proportion of artificial nearest neighbours for varying
# combinations of the pN and pK
sweep_list <- paramSweep(sample, PCs = 1:min_pc, sct = FALSE)
sweep_stats <- summarizeSweep(sweep_list)
bcmvn <- find.pK(sweep_stats) # computes a metric to find the optimal pK value (max mean variance normalised by modality coefficient)
# Optimal pK is the max of the bimodality coefficient (BCmvn) distribution
optimal.pk <- bcmvn %>%
dplyr::filter(BCmetric == max(BCmetric)) %>%
dplyr::select(pK)
optimal.pk <- as.numeric(as.character(optimal.pk[[1]]))Number of expected real doublets, nExp
Doublet detection tools always find it tricky to identify homotypic doublets, that is, doublets that are made up of 2 identical cell states. DoubletFinder offers a solution to this issue, by correcting the multiplet rate to account for this.
First, let’s estimate the homotypic doublet proportion. For this we need to know our cell types – I’m using the clusters as cell types, assuming that cells that were clustered together are the same cell type.
# Homotypic doublet proportion estimate
annotations <- sample@meta.data$seurat_clusters # use the clusters as the user-defined cell types
homotypic.prop <- modelHomotypic(annotations) # get proportions of homotypic doubletsNext, we need the multiplet rate of our sample. This is dependent on the number of cells that was loaded into the sequencer, as well as the platform, etc. You might be able to get this information from the sequencing facility. Otherwise, as an example, these are the published expected multiplet rates when using 10x (which might change if you are using a different chemistry!). If you provide a multiplet rate, this step will be skipped, otherwise, it is taking the number of cells in your seurat object as an approximation of the number of recovered cells, to select the appropriate multiplet rate.
# Get the multiplet rate if not provided
if(is.null(multiplet_rate)){
print('multiplet_rate not provided....... estimating multiplet rate from cells in dataset')
# 10X multiplet rates table
#https://rpubs.com/kenneditodd/doublet_finder_example
multiplet_rates_10x <- data.frame('Multiplet_rate'= c(0.004, 0.008, 0.0160, 0.023, 0.031, 0.039, 0.046, 0.054, 0.061, 0.069, 0.076),
'Loaded_cells' = c(800, 1600, 3200, 4800, 6400, 8000, 9600, 11200, 12800, 14400, 16000),
'Recovered_cells' = c(500, 1000, 2000, 3000, 4000, 5000, 6000, 7000, 8000, 9000, 10000))
print(multiplet_rates_10x)
multiplet_rate <- multiplet_rates_10x %>% dplyr::filter(Recovered_cells < nrow(seu_sample_subset@meta.data)) %>%
dplyr::slice(which.max(Recovered_cells)) %>% # select the min threshold depending on your number of samples
dplyr::select(Multiplet_rate) %>% as.numeric(as.character()) # get the expected multiplet rate for that number of recovered cells
print(paste('Setting multiplet rate to', multiplet_rate))
}
Once we have the multiplet rate, we can multiply it by the number of cells to get the number of expected multiplets. The number of doublets is then calculated with the following formula:
nExp.poi <- round(multiplet_rate * nrow(sample@meta.data)) # multiply by number of cells to get the number of expected multiplets
nExp.poi.adj <- round(nExp.poi * (1 - homotypic.prop)) # expected number of doubletsFinally, run DoubletFinder
Amazing! We got through the toughest bit. Finally, we can run DoubletFinder, really easily, by just filling out the different parameters we computed.
I change the name of the column, to “doublet_finder” but this is not really necessary.
# run DoubletFinder
sample <- doubletFinder(seu = sample,
PCs = 1:min_pc,
pK = optimal.pk, # the neighborhood size used to compute the number of artificial nearest neighbours
nExp = nExp.poi.adj) # number of expected real doublets
# change name of metadata column with Singlet/Doublet information
colnames(sample@meta.data)[grepl('DF.classifications.*', colnames(sample@meta.data))] <- "doublet_finder"
DoubletFinder will add the annotations to your seurat object directly, but I extract them to return a dataframe, so I can decide whether to add it to the metadata or not.
double_finder_res <- sample@meta.data['doublet_finder'] # get the metadata column with singlet, doublet info
double_finder_res <- rownames_to_column(double_finder_res, "row_names") # add the cell IDs as new column to be able to merge correctly
return(double_finder_res)And that’s it!
2. Add DoubletFinder results to Seurat object
When we actually run this function, per sample, with lapply(), we get a list of dataframes for every sample, which we then merge into a single dataframe.
And then, we make sure the rownames are the same as in the Seurat object, so the cell IDs, because we can easily use AddMetadata to add the doublet_finder dataframe to out Seurat object.
sglt_dblt_metadata <- data.frame(bind_rows(samp_split)) # merge to a single dataframe
rownames(sglt_dblt_metadata) <- sglt_dblt_metadata$row_names # assign cell IDs to row names to ensure match
sglt_dblt_metadata$row_names <- NULL
head(sglt_dblt_metadata)
seu <- AddMetaData(seu, sglt_dblt_metadata, col.name = 'doublet_finder')2. Summary of doublet detection results
Nice! Let’s have a look at a violin plot with different QC stats, separating singlets and doublets.
# Check how doublets singlets differ in QC measures per sample.
VlnPlot(seu, group.by = 'SampleID', split.by = "doublet_finder",
features = c("nFeature_RNA", "nCount_RNA", "percent_mt", "percent_ribo", "percent_hb"),
ncol = 3, pt.size = 0) + theme(legend.position = 'right')For example, let’s plots violin plots of quality control metrics (number of RNA features, RNA counts, mitochondrial percentage, ribosomal percentage, hemoglobin percentage) split by `DoubletFinder` class and grouped by `SampleID`. As you can see doublets tend to have a higher number of genes and read counts, but we don’t expect big differences in the mitochondrial percentage or ribosomal percentage.
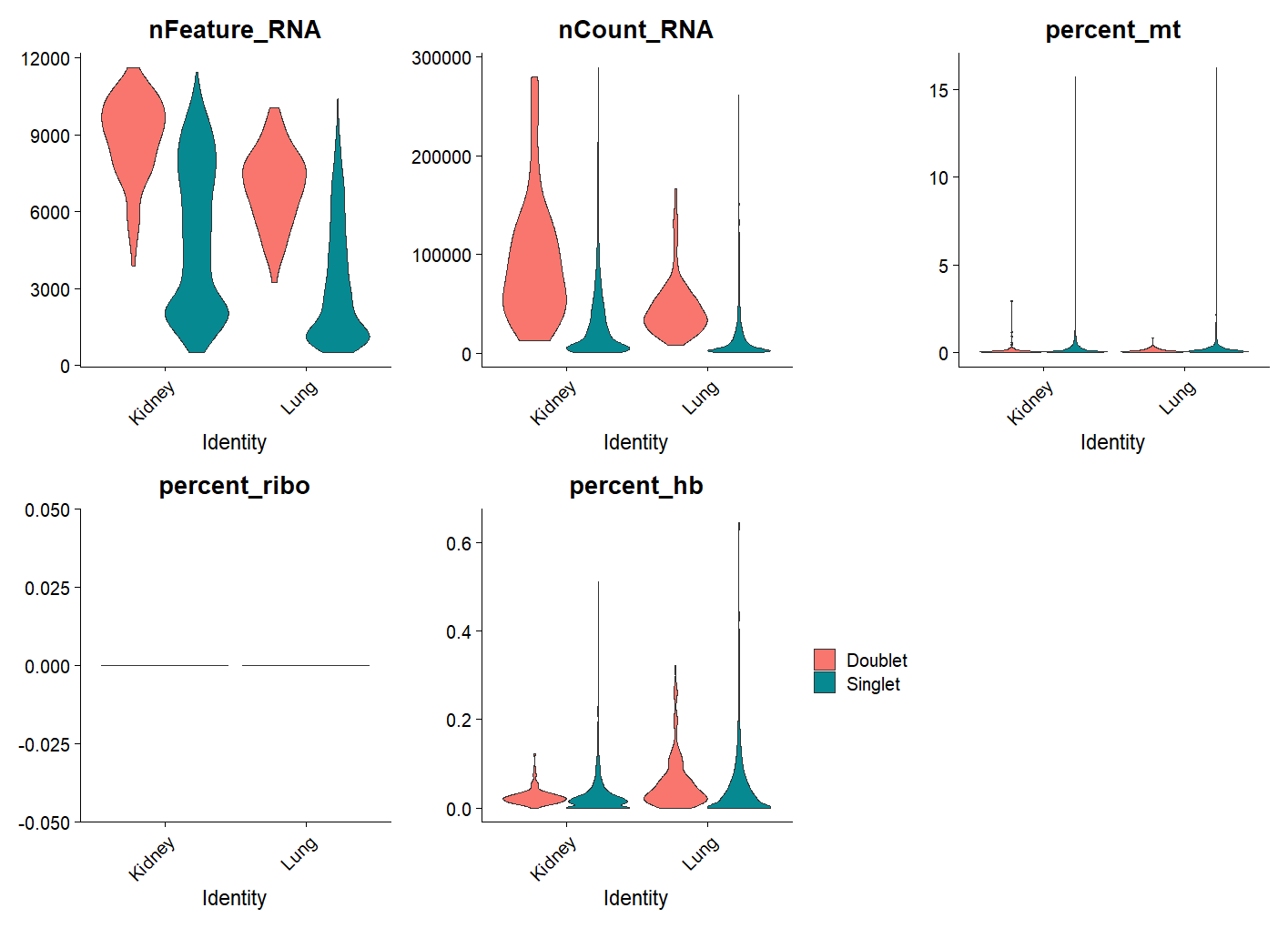
We can also create a nice summary table by grouping the metadata by `SampleID` and `doublet_finder` and calculating the total count of cells in each group and the percentage of doublets and singlets for each sample.
And of course we can save this summary table to a text file.
# Get doublets per sample
doublets_summary <- seu@meta.data %>%
group_by(SampleID, doublet_finder) %>%
summarise(total_count = n(),.groups = 'drop') %>% as.data.frame() %>% ungroup() %>%
group_by(SampleID) %>%
mutate(countT = sum(total_count)) %>%
group_by(doublet_finder, .add = TRUE) %>%
mutate(percent = paste0(round(100 * total_count/countT, 2),'%')) %>%
dplyr::select(-countT)
write.table(doublets_summary, file = file.path(doublet_folder, paste0(proj, '_doubletfinder_doublets_summary.txt')), quote = FALSE, row.names = FALSE, sep = '\t')
> doublets_summary
# A tibble: 4 × 4
# Groups: SampleID, doublet_finder [4]
SampleID doublet_finder total_count percent
<chr> <chr> <int> <chr>
1 Kidney Doublet 93 1.87%
2 Kidney Singlet 4881 98.13%
3 Lung Doublet 140 2.78%
4 Lung Singlet 4896 97.22% 5. Remove doublets
You can remove cells marked as “doublet” very easily with the function subset().
# Removing doublets ============================================================
# If all is ok subset and remove doublets
seu_dblt <- subset(doublet_finder == 'Singlet')And that’s it! We have successfully detected and removed doublets with DoubletFinder in R!
sessionInfo()
Check my sessionInfo() here in case you have trouble reproducing my steps:
> sessionInfo()
R version 4.3.2 (2023-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 11 x64 (build 22631)
Matrix products: default
locale:
[1] LC_COLLATE=English_Ireland.utf8 LC_CTYPE=English_Ireland.utf8 LC_MONETARY=English_Ireland.utf8 LC_NUMERIC=C
[5] LC_TIME=English_Ireland.utf8
time zone: Australia/Sydney
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices datasets utils methods base
other attached packages:
[1] DoubletFinder_2.0.4 scDblFinder_1.12.0 SingleCellExperiment_1.24.0 SummarizedExperiment_1.32.0 Biobase_2.62.0
[6] GenomicRanges_1.54.1 GenomeInfoDb_1.38.5 IRanges_2.36.0 S4Vectors_0.40.2 BiocGenerics_0.48.1
[11] MatrixGenerics_1.14.0 matrixStats_1.2.0 Seurat_5.0.1 SeuratObject_5.0.1 sp_2.1-2
[16] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2
[21] readr_2.1.5 tidyr_1.3.1 tibble_3.2.1 ggplot2_3.4.4 tidyverse_2.0.0 And that is the end of this tutorial!
This is a very simple workflow for detecting, summarizing, and visualizing doublets in scRNA-seq data using Seurat and DoubletFinder in R.
When detecting doublets in scRNAseq data, you often want to test out several tools and even finetune parameters for each tool. Another tool I’ve tried that has given me great results and is relatively easy to use is scDblFinder, for which I have a tutorial here. In this other blogpost, I compare both tools and remove only high-confidence doublets (doublets that were found by both tools).
You can read more abut DoubletFinder here.
Before you go, you might want to check:
- DoubletFinder method – easy explanation: https://www.youtube.com/watch?v=E-0N16CS7MQ&t=3s
- scDblFinder tutorial: https://biostatsquid.com/scdblfinder-tutorial/
- DoubletFinder publication: https://pubmed.ncbi.nlm.nih.gov/30954475/
- DoubletFinder github: https://github.com/chris-mcginnis-ucsf/DoubletFinder
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and… see you in the next one!
Squids don't care much for coffee,
but Laura loves a hot cup in the morning!
If you like my content, you might consider buying me a coffee.
You can also leave a comment or a 'like' in my posts or Youtube channel, knowing that they're helpful really motivates me to keep going:)
Cheers and have a 'squidtastic' day!


Hi!
Thank you for the great tutorial.
I was wondering if you have a typo on the code here in the tutorial (paramSweep_v3 instead of paramSweep). I checked your github for a custom function and couldn’t find it.
Thank you!
Hi! Thanks for noticing and posting! I think since I made this tutorial they updated DoubletFinder (https://github.com/chris-mcginnis-ucsf/DoubletFinder/) but as far as I can see, with the latest version it should be paramSweep. Do let me know if it’s not working for you though!
Hello,
Please can I ask what your would do if the Pk value is NULL?
bcmvn <- find.pK(sweep_stats) # computes a metric to find the optimal pK value (max mean variance normalised by modality coefficient)
NULL
Thanks!
Hi! It’s very tricky to debug without seeing your environment and code, but this is what I would try to debug:
The most likely cause is that the pK column in your sweep_stats contains all NA values or the variance calculations failed.
Try to first check what sweep_stats looks like and if it is as expected (perhaps run it on DoubletFinder’s test dataset first):
str(sweep_stats)
head(sweep_stats)
summary(sweep_stats)
You can also try to regenerate sweep_stats with different parameters.
Try also checking for sufficient cells and variation:
print(ncol(seu_object))
print(range(seu_object@reductions$pca@cell.embeddings[,1:20]))
Good luck!
Hello!
Thank you for this great tutorial. I just had a doubt about whether to do doublet removal before or after integration? Is the correct workflow then to do QC, SCTransform, integrate, run umap, pca, clustering, then doublet removal? I saw many people doing doublet removal after QC too?
Thank you for all your amazing work.
Hey! Thanks so much for your comment. In most workflows I’ve seen, doublet removal should be done after QC but before integration. This makes sense as you do not want doublets to confound integration algorithms. Also, after integration, the data has been corrected for batch effects and aligned across samples. That’s great for clustering, but bad for doublet detection — because doublets may look like “intermediate” cells between clusters, and integration can “pull” those toward one group. So doublets become harder to identify post-integration.
So if you detect doublets after integration, you risk:
– Missing many real doublets (false negatives)
– Removing rare or transitional cell types incorrectly (false positives)
Not sure in which cases you saw it after integration, but I’m keen to know why! So if you’d like you can share those examples:)
Hi, thanks for the tutorial! in a multi-sample seurat object, do you run everything as one seurat obj up until the custom doublet finder function? Or should all the preprocessing (PCA, UMAP, pK, multiplet rate) be done on the list of separated seurat objects?
Hi! Really good question – depends a bit on your experiment design but it’s probably best to run preprocessing (filtering + doublet detection) per sample, as some samples may be noisier than others, and their libraries would have been prepared separately even if they are multiplexed. Would definitely run DoubletFinder per sample because it relies on calculating a pK value (the neighborhood size). This value is highly sensitive to the unique noise profile, cell density, and library quality of an individual sequencing run. But you can still do that within a single Seurat object (that’s one of the main points of Seurat v5’s layers!).
So general steps would be:
1. Create one object: Merge all your samples into one Seurat object.
2. Keep Layers Split: Use obj[[“RNA”]] <- split(obj[["RNA"]], f = obj$sample_id). This keeps the counts separate. 3. Loop for Doublets: Use a lapply or a simple for loop to run NormalizeData, FindVariableFeatures, ScaleData, RunPCA, and DoubletFinder on each sample layer individually. (and filtering of low quality cells before that if you need to) 4. Filter: Remove the predicted doublets from the main object. 5. Re-Join & Integrate: Only after the doublets are gone do you join the layers (or use an integration tool like Harmony or CCA) to run your final, global UMAP.