A short but simple explanation of t-SNE – easily explained with an example!
PCA, t-SNE, UMAP… you’ve probably heard about all these dimensionality reduction methods. In this series of blogposts, we’ll cover the similarities and differences between them, easily explained!
In this post, you will find out what is t-SNE and how to interpret it with an example.
You can check part 1: easy PCA here.
So if you are ready… let’s dive in!
Click on the video to follow my easy tSNE explanation with an example on Youtube!
What is dimensionality reduction?
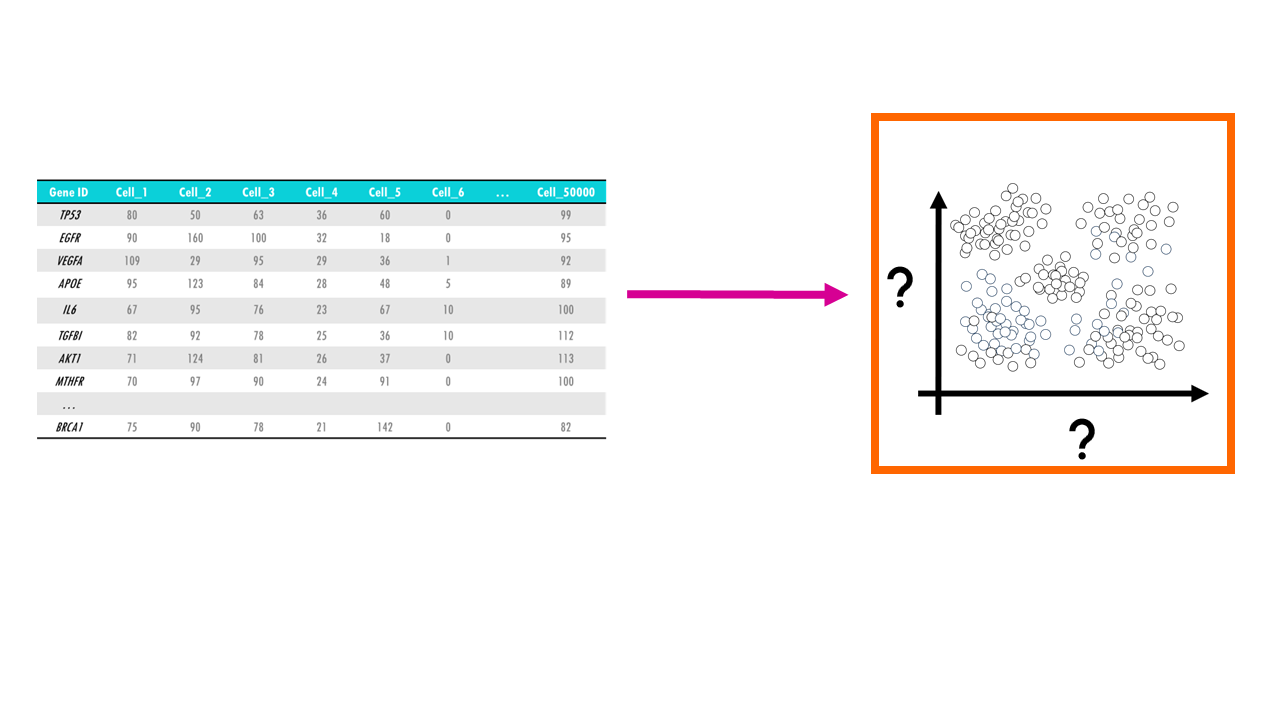
Let’s imagine you have a single-cell dataset with the gene expression of 10000 genes across 50.000 cells. For each cell, and for each gene, you have a gene expression value that tells you how much that gene was expressed in that cell.
Sometimes, we just want to visualise large datasets like this one in a plot. If each point in the plot is a cell, that would allow us to identify clusters of cells with similar expression levels of certain genes, we can identify cell types, or cells that are very different to the rest in their gene expression profile. The problem with such big datasets like this one, is that we cannot visualise all features, or genes, at once. A plot will have two axes, three at most. So we could pick two genes, and plot each cell according to their expression level of MCM3 and AKT1. We could then identify clusters of cells which have a high expression of both MCM3 and AKT1, and cells here have high expression of MCM3 but relatively low expression of AKT1, for example. However, this plot leaves out the expression level of the other 9998 genes in our dataset.
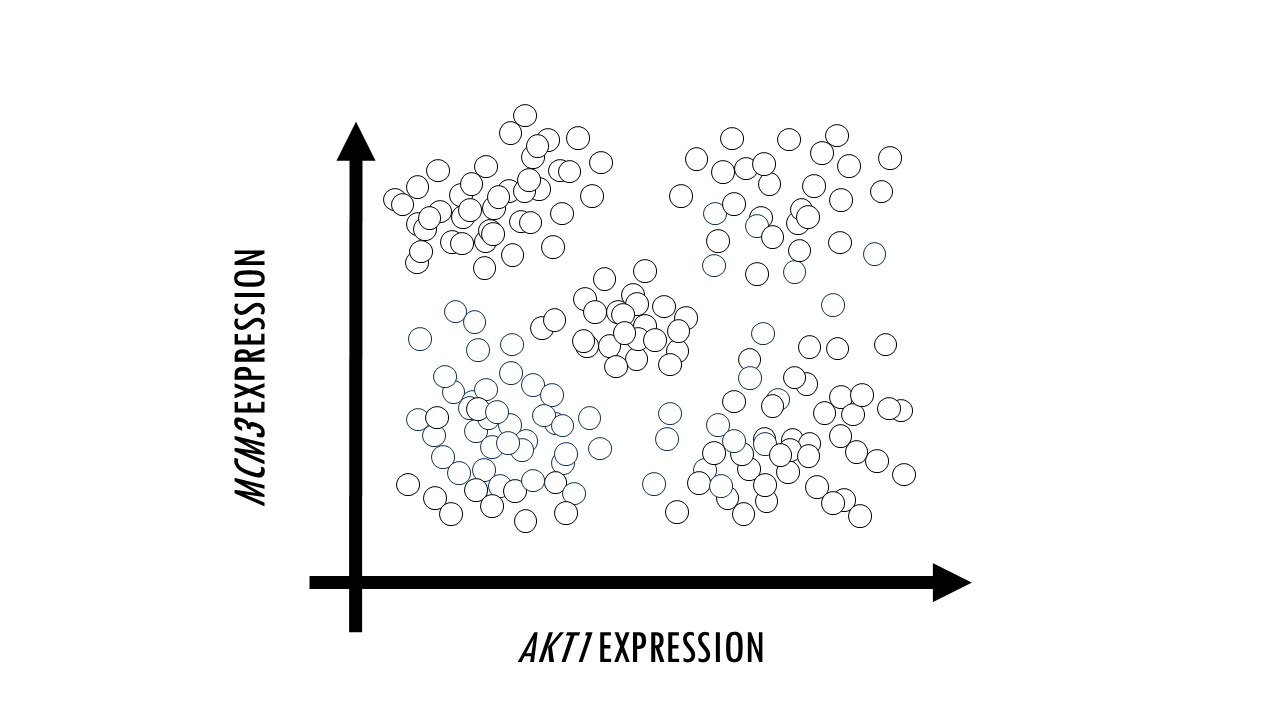
In other words, we need a way to convert a multidimensional dataset into two variables we can plot. This process is called dimensionality reduction and through it we basically reduce the number of input variables or features (in this case, genes) while retaining maximum information. Obviously, when we reduce the number of dimensions, we are going to lose information, but the idea is to preserve as much of the dataset’s structure and characteristics as possible.
To do this, we need a dimension reduction algorithm, which is basically a method that reduces the number of dimensions. As you can imagine, there are many dimension reduction algorithms out there, but we are going to talk about 3 of the most common ones: PCA, UMAP and t-SNE. We will go through the main ideas of each of them and the differences between them, so you can decide which one is most appropriate for your data.
Squidtip
Dimensionality reduction is a process that transforms data from high-dimensional space into a low-dimensional space while preserving as much of the dataset’s original information as possible.
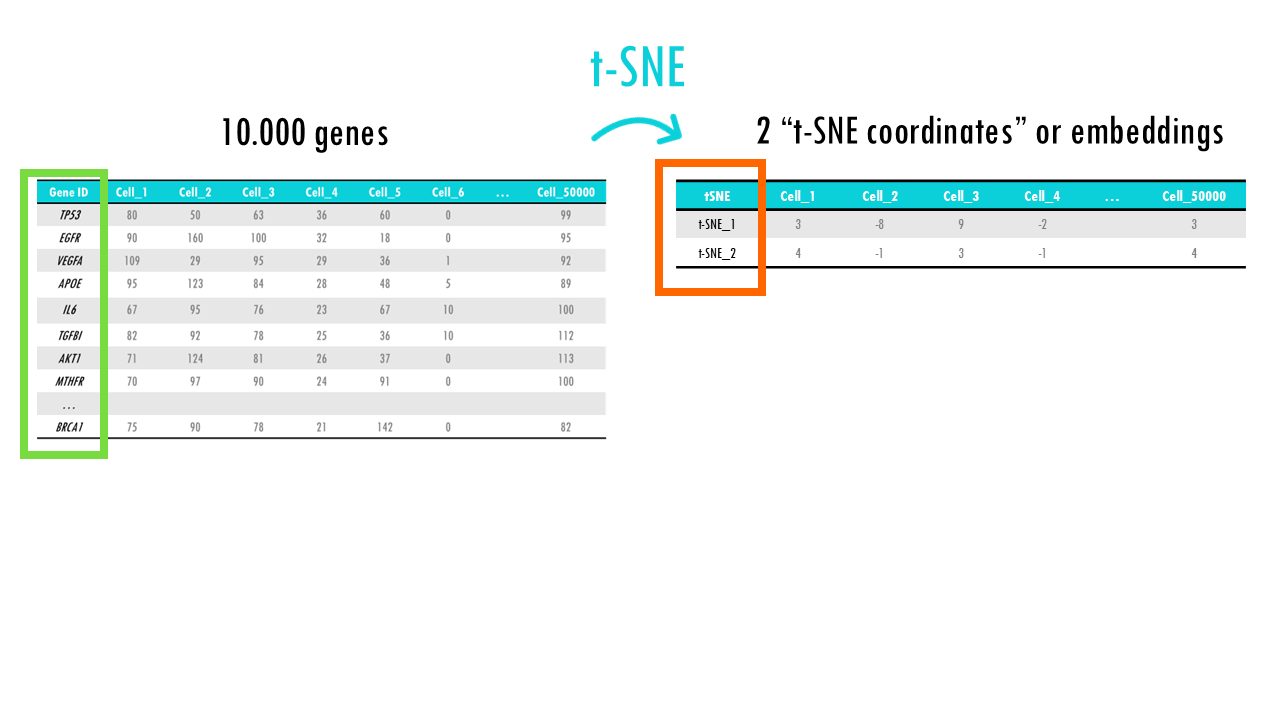
t-distributed Stochastic Neighbour Embedding (t-SNE)
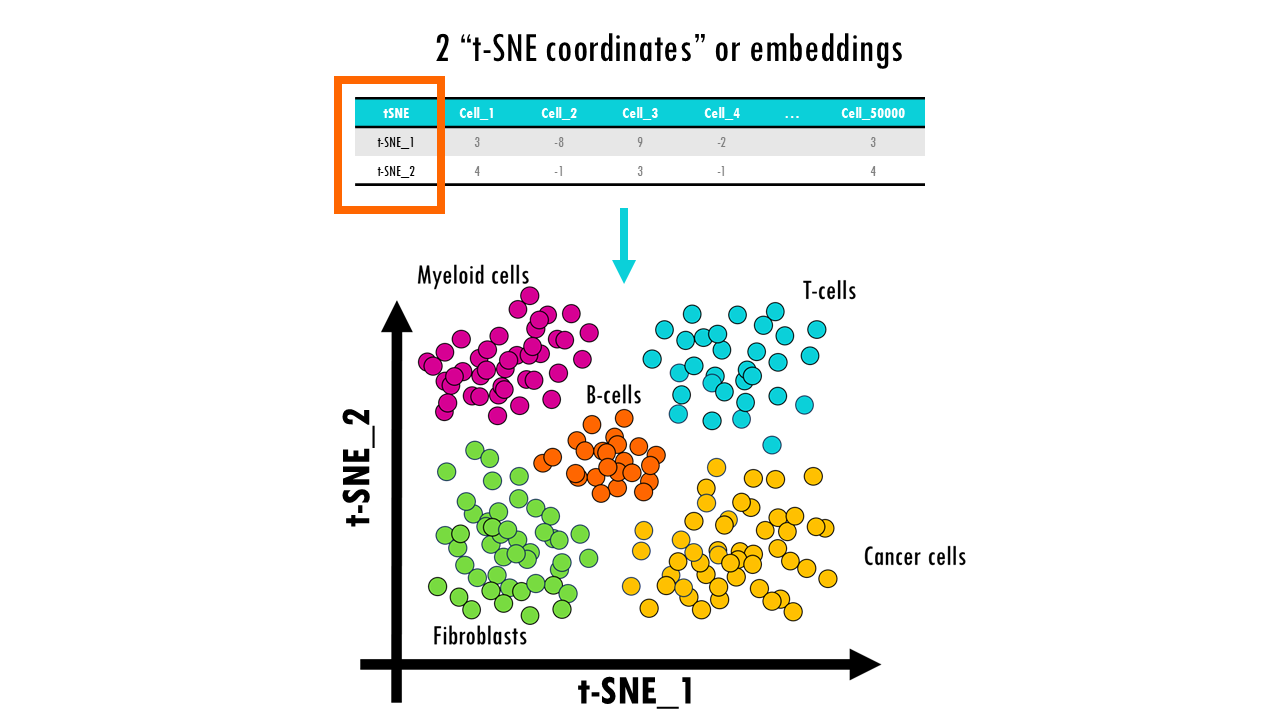
In essence, t-SNE transforms each high-dimensional object (in this case, each cell which has many genes) to a two-dimensional point (sometimes three), in such a way, that when we plot it, cells with similar gene profiles will be assigned to nearby points, and cells with very different gene profiles will have distant coordinates. In other words, t-SNE will keep similar cells close together, and push cells that are different further apart. Great! That’s exactly what we want!
Squidtip
t-SNE is a non-linear dimensionality reduction algorithm particularly well-suited for visualizing high-dimensional data by modelling each high-dimensional object by a two- or three-dimensional point in such a way that similar objects are modelled by nearby points and dissimilar objects are modelled by distant points.
How does t-SNE work?
So how does t-SNE do that? I’ll give you a very high-level explanation of the maths involved in this transformation. If you are interested in a more rigorous, math-sy explanation, check out the links below!
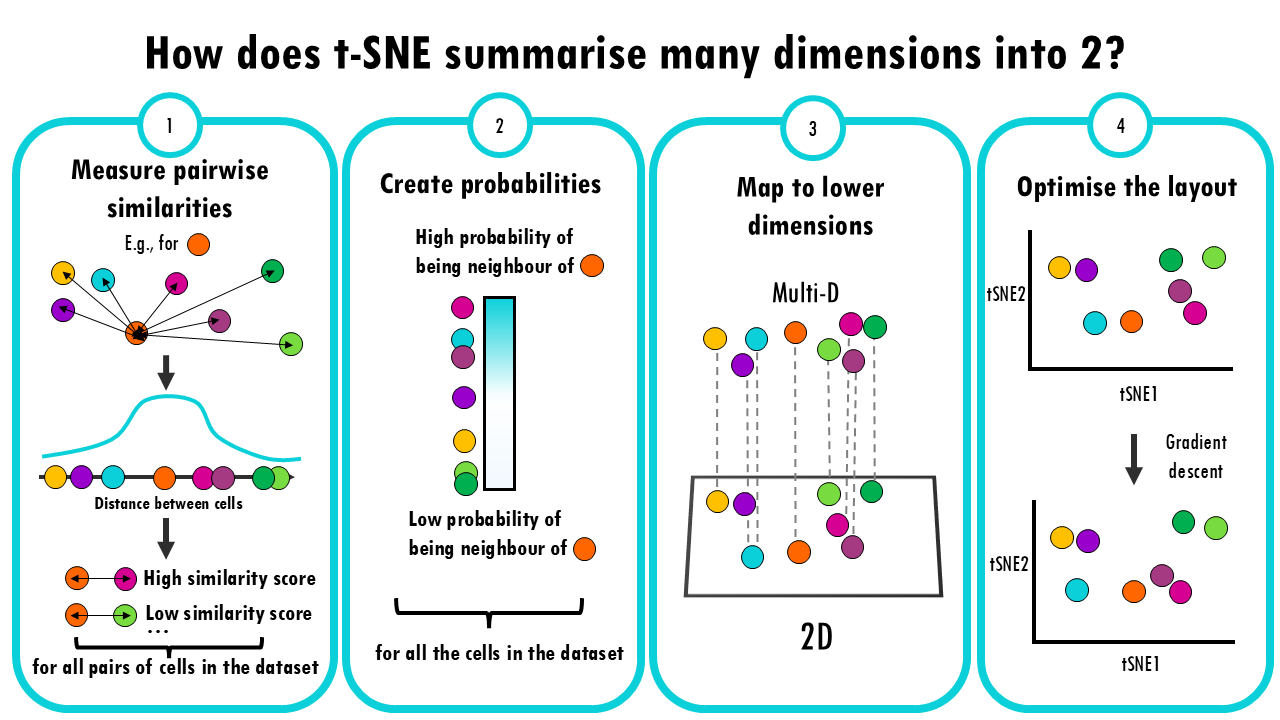
- Measure pairwise similarities: First, t-SNE calculates how similar each pair of cells is to each other. It does this by looking at the “distance” between them, often using a method like Gaussian (normal) distribution. The idea is that if two cells have very similar gene expression profiles, they should have a high similarity score, and if they’re far apart, the similarity should be low.
- Create probabilities: These similarities are turned into probabilities (think of it like a “likelihood” that two cells are close neighbours). The closer two points are, the higher the probability that they are neighbours.
- Map to lower dimensions: Now, t-SNE creates a new 2D space and tries to position the data points there. The goal is to place points so that similar cells in the original space are still close together in the new space, and dissimilar cells are far apart.
- Optimize the layout: This is where the “stochastic” part comes in. t-SNE uses a technique called gradient descent, which is a way of adjusting the positions of the points in the lower-dimensional space step by step, trying to make the distribution of similarities in the lower space match the original distribution as closely as possible.
Squidtip
How does t-SNE “optimise the layout”?
In step 4; t-SNE computes two distributions: a distribution that measures pairwise similarities of the input objects and a distribution that measures pairwise similarities of the corresponding low-dimensional points. It then tries to minimize the divergence between two distributions.
The key math involves probabilities and optimization, but the goal is to create a map that reflects the relationships between the cells as closely as possible, just in fewer dimensions.
In summary, t-SNE is excellent for visualizing complex, high-dimensional data in two or three dimensions (for example, if we want to see clusters of cells with similar gene expression profiles). But, it is computationally expensive and sensitive to hyperparameters, meaning the plot you get at the end depends on the perplexity you set.
t-SNE hyperparameters
Wait a minute, hyperparameters?
The main hyperparameter I am taking about is the perplexity, which controls how the algorithm balances local versus global relationships in the data. Perplexity is related to how many neighbours each point is considering when building its similarity map.
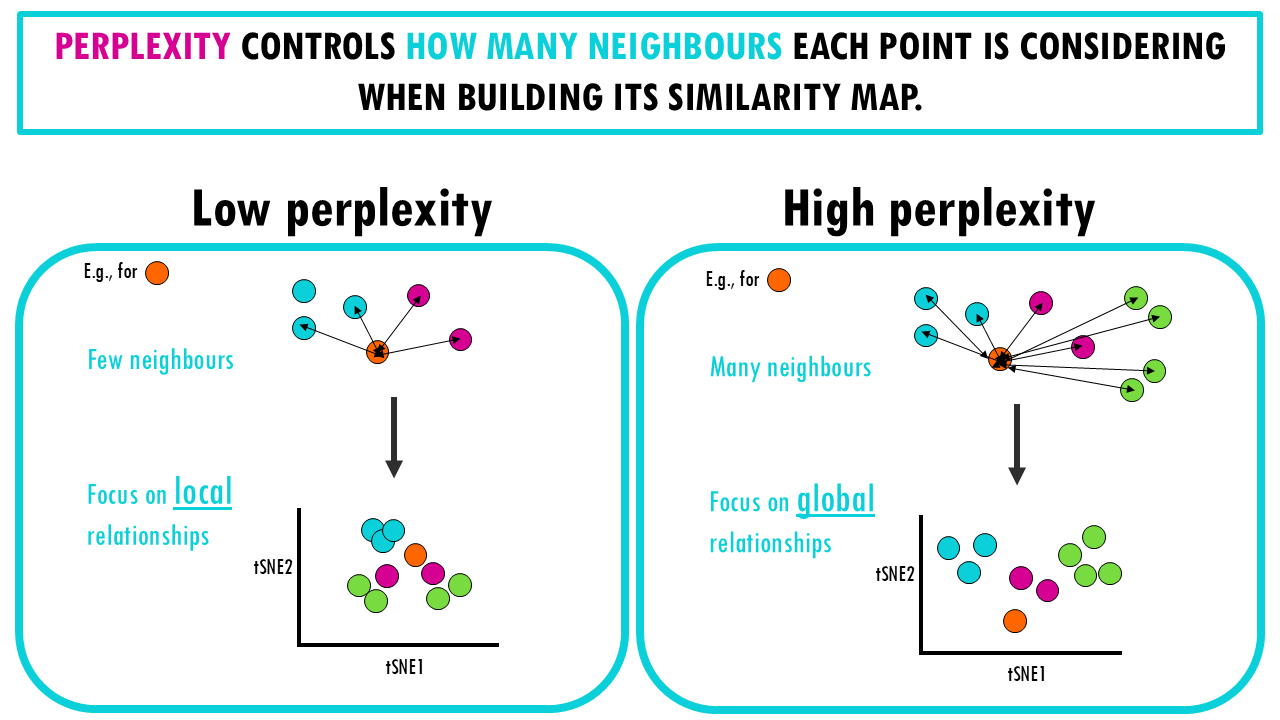
- When we set perplexity to a low value, we only consider a few neighbours when checking similarity, so the algorithm focuses more on local relationships. This means it will be sensitive to small differences between cells and will try to preserve tight clusters of cells that have very similar gene expression profiles.
- When you set perplexity to a high value, the algorithm starts to consider a larger number of neighbouring cells when calculating similarity. This makes it better at capturing global relationships, like how different cell types (e.g., neurons, epithelial cells, immune cells) are distributed across the dataset. The individual clusters of cells might be less tightly packed, but the overall arrangement of cell types is probably clearer. The immune cells might form a cluster, and the epithelial cells might form another, even if there is some variability in gene expression within each group.
Squidtip
What’s the maths behind the perplexity hyperparameter?
When t-SNE calculates how similar points are to each other, it uses a Gaussian distribution to model the likelihood that two points are neighbours. Perplexity helps determine the “spread” of that Gaussian. A lower perplexity means we are considering only a small number of neighbours when building the similarity map, making the method focus more on local structure (tight, small groups of similar points). A higher perplexity means t-SNE will consider a broader range of neighbours, capturing more of the global structure (larger, more spread-out relationships).
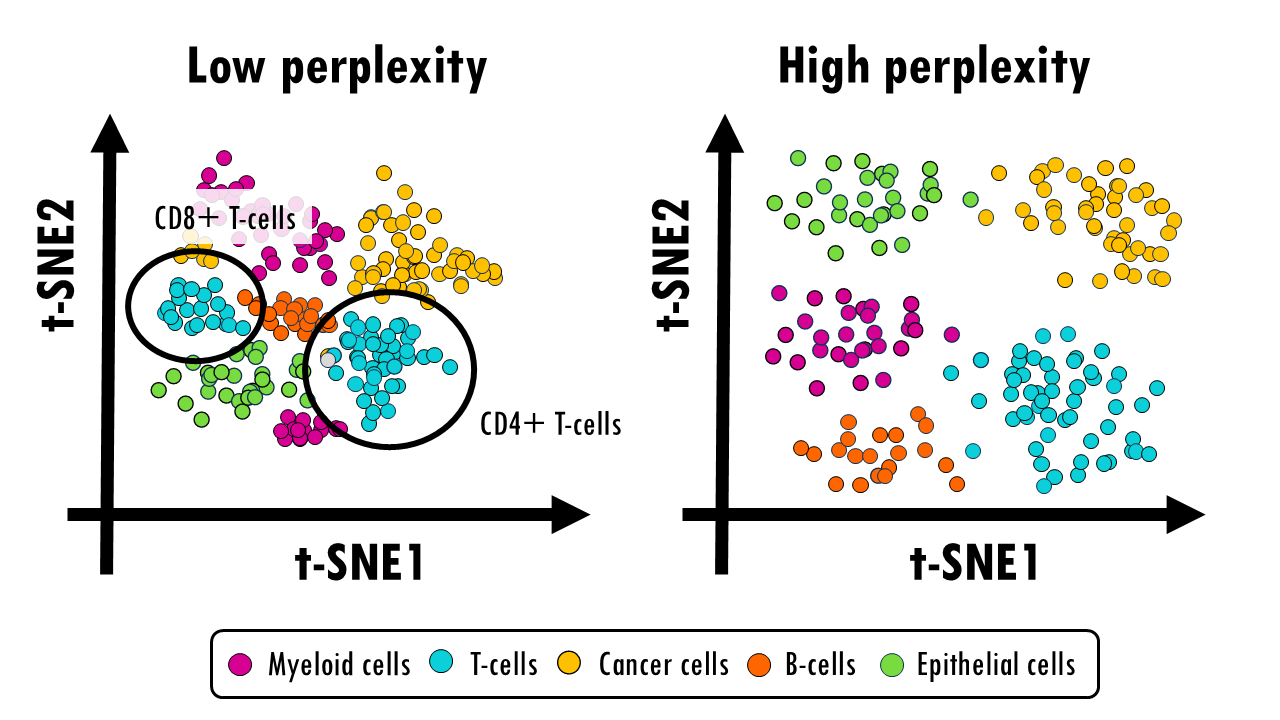
In our scRNAseq data, setting a low perplexity would help us visualize small differences between immune cell subtypes (e.g., T CD4+ cells and T CD8+ cells). But it might not preserve the large-scale organization between cell types like immune cells vs. non-immune cells, and these might appear more mixed.

Final notes on t-SNE
In summary, t-SNE is a dimensionality reduction algorithm which allows us to visualise high-dimensional data in a 2D plot. It does it by modelling each high-dimensional object by a two- or three-dimensional point in such a way that similar objects are modelled by nearby points and dissimilar objects are modelled by distant points.
However, t-SNE also has limitations: it is computationally expensive (takes really long with very large datasets!) and sensitive to its main hyperparameter: perplexity. So t-SNE does require a bit of optimisation to find the right perplexity to visualise your dataset.
In part 3 of this series, we’ll talk about another similar dimensionality reduction algorithm: UMAP!
Want to know more?
Additional resources
If you would like to know more about t-SNE, check out:
- t-SNE: a more in-depth math explanation (Towards Data Science)
- t-SNE: step-by-step of the maths behind it!
You might be interested in…
- UMAPs simply explained
- PCA simply explained
Ending notes
Wohoo! You made it ’til the end!
In this post, I shared some insights into t-SNE and how it works.
Hopefully you found some of my notes and resources useful! Don’t hesitate to leave a comment if there is anything unclear, that you would like explained further, or if you’re looking for more resources on biostatistics! Your feedback is really appreciated and it helps me create more useful content:)
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and… see you in the next one!
Squids don't care much for coffee,
but Laura loves a hot cup in the morning!
If you like my content, you might consider buying me a coffee.
You can also leave a comment or a 'like' in my posts or Youtube channel, knowing that they're helpful really motivates me to keep going:)
Cheers and have a 'squidtastic' day!
And that is the end of this tutorial!
In this post, I explained the differences between log2FC and p-value, and why in differential gene expression analysis we don't always get both high log2FC and low p-value. Hope you found it useful!
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and... see you in the next one!


Hi there … i really like how simplified and attractive your post about t-SNE was and i was so settled and interested how you beautifully explained it using such interesting colors and diagrams tysmmm ….
Thank you so much for your comment! So glad you liked it:)