A step-by-step easy R tutorial to detect and remove doublets from scRNAseq data
In this easy, step-by-step tutorial you will learn how to do some general doublet QC and remove doublets from our Seurat object in R for scRNAseq data.
This is part 3 of my tutorial series on doublet detection and assumes you have already run 1 or more tools for doublet detection so you have your singlet and doublet annotations in the metadata. So if you haven’t yet, check out my part 1 and 2 tutorials on doublet detection using scDbltFinder, and DoubletFinder to identify doublets.
However, there are many tools to infer doublets from scRNAseq data, and you can also use code snippets from this tutorial if you used other doublet detection tools. You can read more about other doublet detection tools in this benchmarking paper here.
As you probably already know, doublets are artifacts that occur when two cells are captured together and sequenced as one. This means that in your matrix, for a given cell barcode, you are getting gene expression data from 2 different cells. These doublets can really confound your downstream analysis, so it is important to identify them during the preprocessing steps and remove them.
In this tutorial, we will learn how to:
- compare doublet/singlet labels between two doublet detection tools
- compare some basic single-cell stats like number of genes, % mitochondrial genes…, between doublets and singlets
- remove doublets from your dataset
We will use the R packages:
- tidyverse
- Seurat
If you are more of a video-based learner, you can check out my Youtube video on how to remove doublets from scRNAseq data in R. Otherwise, just keep reading! And remember you can just copy the code by clicking on the top right corner of the code snippets in this post.
Let’s dive in!
Check out my Youtube tutorial to on how to remove doublets from scRNAseq data in R!
Before you start…
In this tutorial, we will pick up where we left off. I am going to be using a Seurat object on which I ran two doublet detection tools: scDblFinder and DoubletFinder. But you may have run different tools for doublet detection, that’s ok. Basically, in the metadata of your Seurat object, you have one or more columns with labels singlet or doublet, for each cell.
As always, you can find the code I am using in this tutorial at biotatsquid’s github page.
Summary of doublets
We can get a quick summary of how many doublets were called by doublet_finder, and how many were called by scDblFinder.
What is the percentage over the total number of cells that was classified as doublets by each tool? As you can see DoubletFinder finds fewer doublets than scDblfinder. Careful! We will talk about this later on, but this could mean I may need to finetune the parameters when I ran each tool. However, when you run different tools you cannot expect a perfect match since their algorithms work differently.
# Doublet QC: scDblFinder vs DoubletFinder ==========================
# Percentage of doublets per sample
seu@meta.data %>% group_by(SampleID) %>% summarise(percent_doublet_doubletfinder = mean(doublet_finder == 'Doublet') * 100,
percent_doublet_scdblfinder = mean(scDblFinder.class == 'doublet') * 100)
# A tibble: 2 × 3
SampleID percent_doublet_doubletfinder percent_doublet_scdblfinder
<chr> <dbl> <dbl>
1 Kidney 1.87 9.01
2 Lung 2.78 9.29If you run a few doublet detection tools, it’s sometimes useful to know how many cells were classified as doublets by both tools, how many were consensus singlets, and how many differ. This is very easy to do with the function table.
table('scDblFinder' = seu$scDblFinder.class, 'DoubletFinder' = seu$doublet_finder)
Get QC on doublets and singlets
Now let’s create a new column, doublet_consensus, which basically marks cells as:
- “singlet” if they were classified as a singlet by both tools
- “doublet“, if both tools agreed and classified it as doublet
- “DFsinglet_SCDBLdoublet“, if it was marked as a singlet by DoubletFinder but as a doublet by scDblFinder doublet
- “DFdoublet_SCDBLsinglet“, if it was marked as a doublet by DoubletFinder but as a singlet by scDblFinder doublet
This is kind of the same thing we were doing with the table function, except now we are labelling each cell. We do this to have a look at some general QC stats, which may help us decide how to filter out our doublets from our dataset.
meta <- seu@meta.data
meta <- meta %>% mutate(doublet_consensus =
ifelse(doublet_finder == 'Singlet' & scDblFinder.class == 'singlet', 'singlet',
ifelse(doublet_finder == 'Doublet' & scDblFinder.class == 'doublet', 'doublet',
ifelse(doublet_finder == 'Singlet' & scDblFinder.class == 'doublet', 'DFsinglet_SCDBLdoublet', 'DFdoublet_SCDBLsinglet'))))Now, let’s get some QC stats and plots.
For example, one of the main QC checks is to see how many doublets were detected per sample. You want to check that not all doublets are coming from one sample.
table(meta$doublet_consensus, meta$SampleID)We see cells that were classified as singlets by both tools tend to have a lower number of features and counts with a violin plot. We can also plot a density plot.
ggplot(meta, aes(x = doublet_consensus, y = nFeature_RNA, fill = doublet_consensus)) +
geom_violin() +
ylab('Number of genes/cell') +
facet_wrap(~SampleID) +
theme_bw()
ggplot(meta, aes(x = doublet_consensus, y = nCount_RNA, fill = doublet_consensus)) +
geom_violin() +
ylab('Number of molecules/cell') +
facet_wrap(~SampleID)
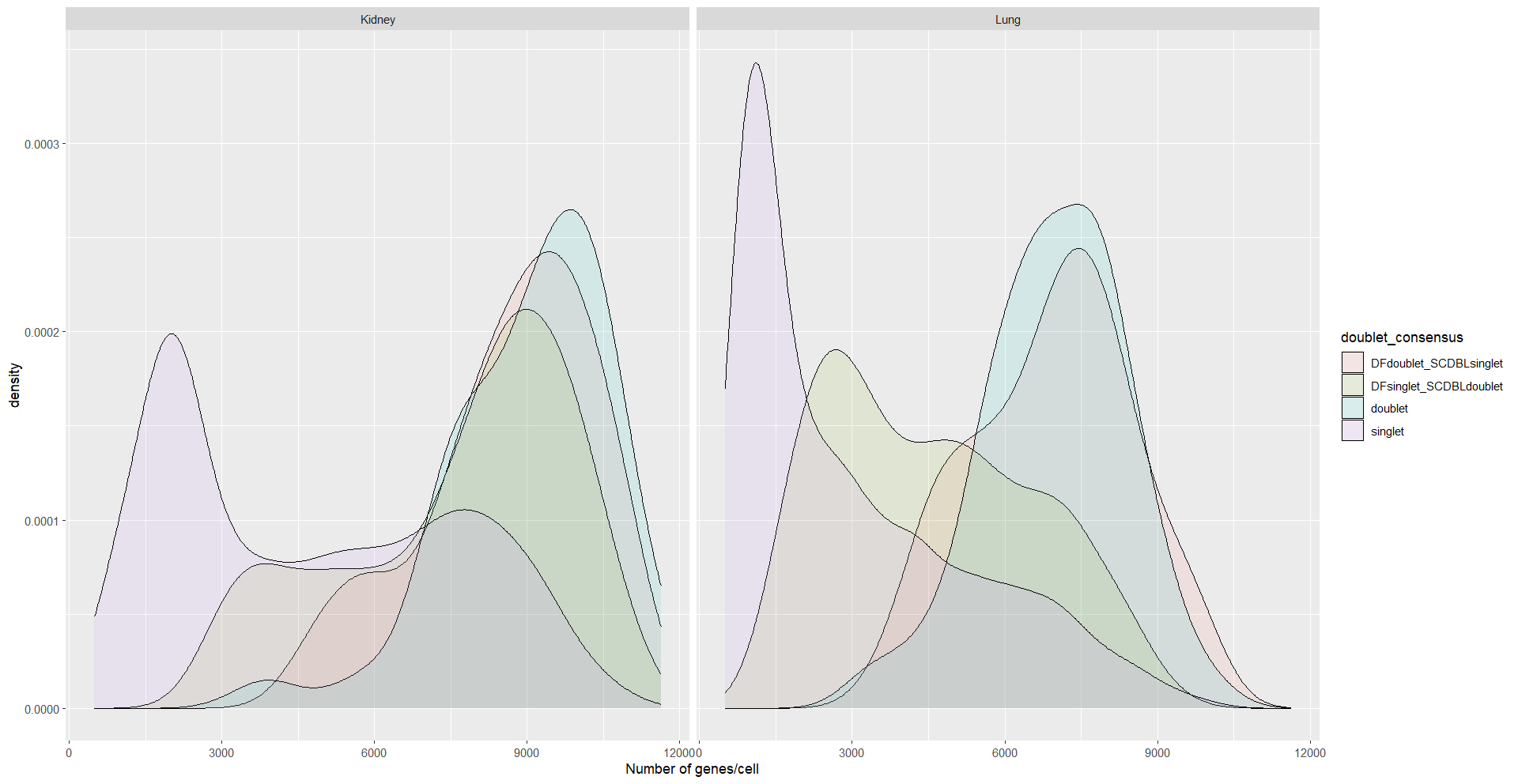
ggplot(meta, aes(x = nFeature_RNA, fill = doublet_consensus)) +
geom_density(alpha = 0.1) +
xlab('Number of genes/cell') +
facet_wrap(~SampleID)
ggplot(meta, aes(x = nCount_RNA, fill = doublet_consensus)) +
geom_density(alpha = 0.1) +
xlab('Number of molecules/cell') +
facet_wrap(~SampleID)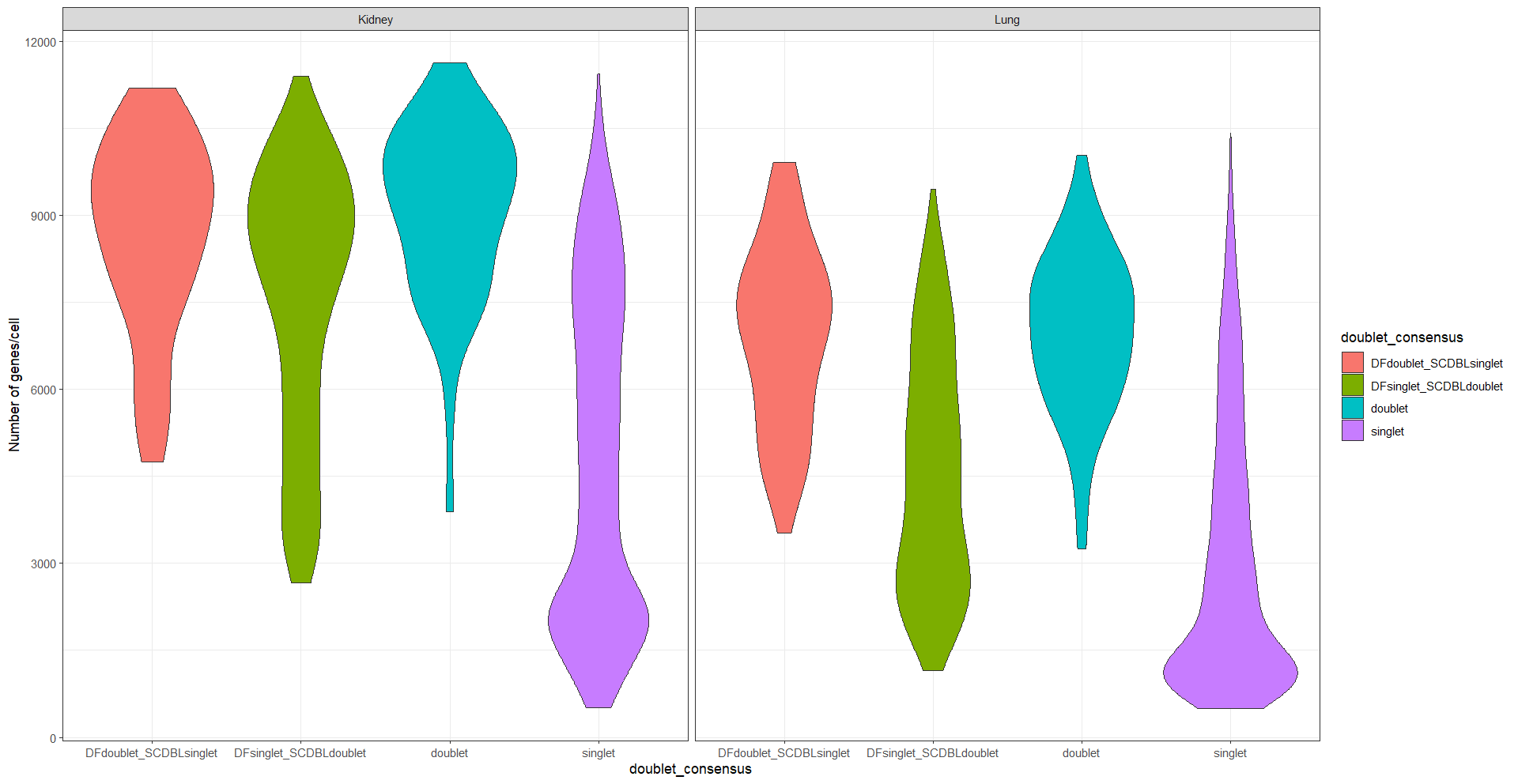
Let’s save our Seurat object with the doublet/singlet annotations, if you haven’t done it yet. I like to save it as an intermediary object before I filter doublet out in case I change my mind later on after downstream analyses and decide to be more or less stringent with my doublet filtering.
# Save seurat with doublet annotation
saveRDS(seu, file = file.path(in_path, 'seu_dblt.rds'))Remove doublets from scRNAseq data
Once you have checked some general QC stats. it’s time to remove doublets from our dataset. We can do this really easily with the function subset(), which is part of the Seurat package, to just keep cells that are marked as singlet. However, you may have already identified an issue here.
We clearly want to remove cells that were classified as doublets by both tools. We also know that we want to keep cells that were classified as singlets by both tools.
But what about cells that were classified as singlets by one tool and doublet by another?
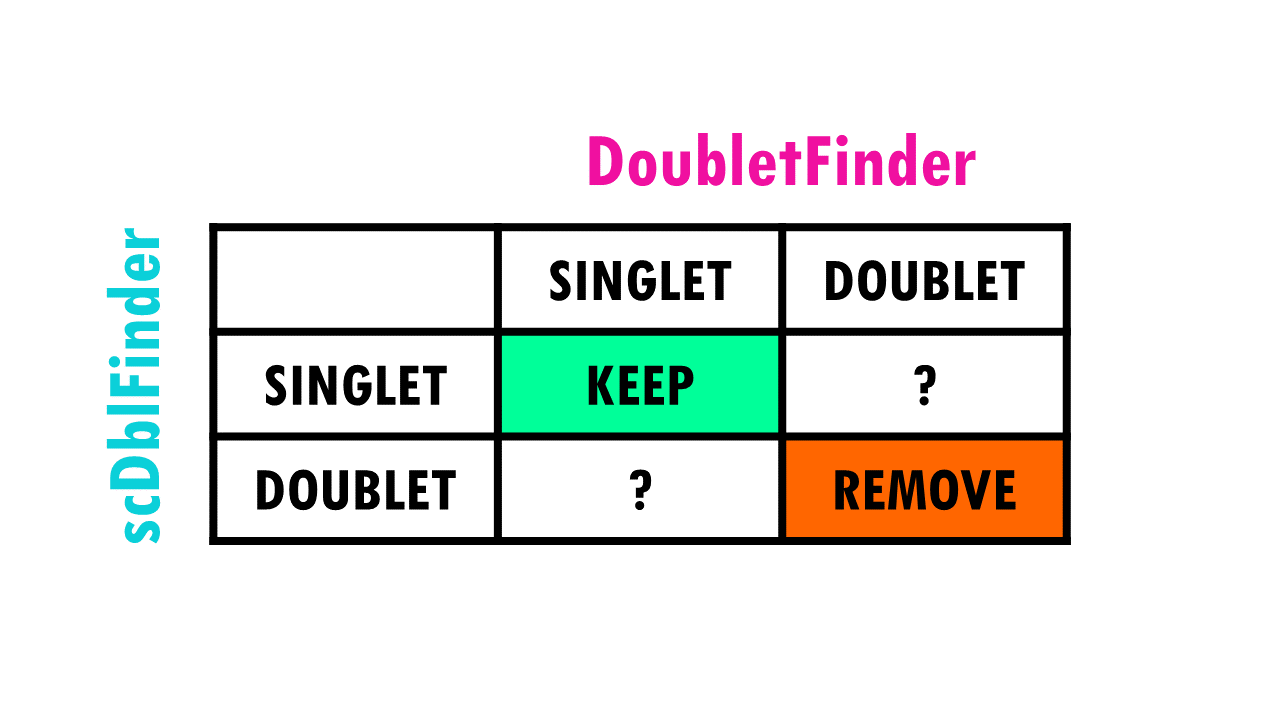
In our case, we have:
- 133 cells that were marked as a “doublet” by DoubletFinder, but “singlet” by scDblFinder.
- 816 cells that were marked as a “doublet” by scDblFinder, but “singlet” by DoubletFinder.
Which tool is correct? They are both inferring if the cell is a doublet from gene expression data, so it is not so easy to decide when there is not a consensus.
There are several things that can help you decide:
- There are some benchmarking papers that compare different doublet finding tools by running them on a dataset with known ground truth and give you stats on the accuracy, so that might help you trust one tool more than the other.
- You can also run a third doublet finding tool, and get the consensus of the 3. More doublet-detection tools here.
- You can also decide to be more lenient, so if at least one tool says it’s a singlet, you keep that cell in. Or be ore stringent, meaning if at least one tool classifies a cell as a doublet, you remove it. It also depends on how many cells you have in your dataset and how many are you willing to remove in this step.
There’s not really a right answer, it also depends on your dataset, and the type of analysis that you are doing! You might proceed with your analysis and get a UMAP of cell clusters, and you might realise you have a cluster of doublets, in which case it’s often advisable to remove that cluster and re-process the data to get cleaner clusters.

In this case, I’m just going to keep cells that were classified as singlets by either tools, because it is a small dataset and I want to “lose” as little cells as possible. (However, I might figure out later on in my analysis that I having a smaller but cleaner dataset is better!)
# Removing doublets ============================================================
# If all is ok subset and remove doublets
# seu_dblt <- readRDS(file.path(in_path, 'seu_dblt.rds'))
seu_dblt <- subset(seu,
subset = doublet_finder == 'Singlet' | scDblFinder.class == 'singlet')
table('scDblFinder' = seu_dblt$scDblFinder.class, 'DoubletFinder' = seu_dblt$doublet_finder)
stopifnot('Something went wrong, genes have been removed!' = dim(seu)[[1]] - dim(seu_dblt)[[1]] == 0)
cat('Total doublets removed: ', dim(seu)[[2]] - dim(seu_dblt)[[2]]) # removed 3500 doublets in total
saveRDS(seu_dblt, file = file.path(in_path, 'seu_dblt_filt.rds'))Nice! It’s always good to doublet check that no genes have been removed, just cells, and you can also get the number of cells you removed.
Finally, we can save the filtered Seurat object.
And that’s it! We have successfully detected and removed doublets from scRNAseq data in R!
sessionInfo()
Check my sessionInfo() here in case you have trouble reproducing my steps:
> sessionInfo()
R version 4.3.2 (2023-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 11 x64 (build 22631)
Matrix products: default
locale:
[1] LC_COLLATE=English_Ireland.utf8 LC_CTYPE=English_Ireland.utf8 LC_MONETARY=English_Ireland.utf8 LC_NUMERIC=C
[5] LC_TIME=English_Ireland.utf8
time zone: Australia/Sydney
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices datasets utils methods base
other attached packages:
[1] DoubletFinder_2.0.4 scDblFinder_1.12.0 SingleCellExperiment_1.24.0 SummarizedExperiment_1.32.0 Biobase_2.62.0
[6] GenomicRanges_1.54.1 GenomeInfoDb_1.38.5 IRanges_2.36.0 S4Vectors_0.40.2 BiocGenerics_0.48.1
[11] MatrixGenerics_1.14.0 matrixStats_1.2.0 Seurat_5.0.1 SeuratObject_5.0.1 sp_2.1-2
[16] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2
[21] readr_2.1.5 tidyr_1.3.1 tibble_3.2.1 ggplot2_3.4.4 tidyverse_2.0.0 And that is the end of this tutorial!
This is a very simple workflow for removing doublets from scRNA-seq data in R.
Before you go, you might want to check:
- scDblFinder tutorial in R: https://www.youtube.com/watch?v=ReXr4F1hhlo
- DoubletFinder tutorial in R: https://www.youtube.com/watch?v=ReXr4F1hhlo&t=1s
- DoubletFinder publication: https://pubmed.ncbi.nlm.nih.gov/30954475/
- DoubletFinder github: https://github.com/chris-mcginnis-ucsf/DoubletFinder
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and… see you in the next one!
Squids don't care much for coffee,
but Laura loves a hot cup in the morning!
If you like my content, you might consider buying me a coffee.
You can also leave a comment or a 'like' in my posts or Youtube channel, knowing that they're helpful really motivates me to keep going:)
Cheers and have a 'squidtastic' day!


0 Comments