BiostatLEARN
Simple and clear explanations of biostatistics methods, statistical concepts and more!
I try to keep them maths-free and straight to the point, with many examples of biological applications.
Latest posts
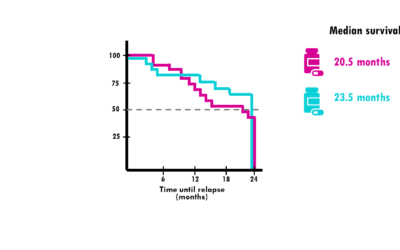
Kaplan-Meier curve – easily explained!
A simple explanation of Kaplan-Meier curves and how to interpret them!In my previous blogpost, we talked about survival time analysis. In a nutshell, survival time analysis, is a group of statistical methods we use to investigate the time it takes for an event of...
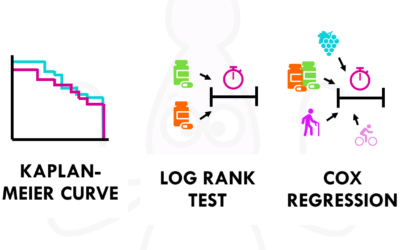
Survival time analysis: easily explained!
A simple introduction to survival time analysis, Kaplan-Meier curves, Cox regression and more!Survival time analysis is really common in biostatistics. You might have heard of Kaplan-Meier curves, Cox regressions or the log rank test. In clinical trials, survival...
Easy confidence intervals and p-values
Confidence intervals, confidence level and p-values simply explained!Let's talk about p-values and confidence intervals! They're very common statistical terms, and I thought I had a clear understanding of what both are, but turns out that language can be misleading...
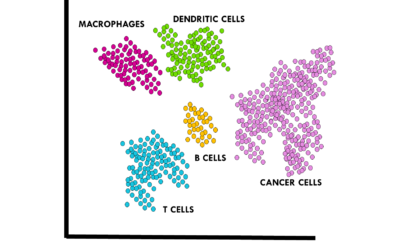
Cell type annotation for scRNAseq
Top tips and resources to perform cell type annotation on scRNAseq dataOnce you preprocess your single-cell RNA sequencing (scRNAseq) data, it is time for one of the biggest challenges in a standard scRNAseq pipeline: annotating cell types. The scientific community...
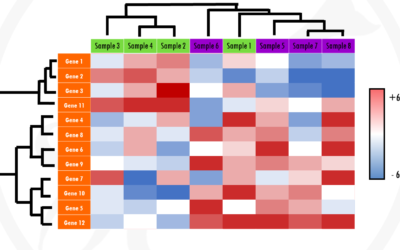
Heatmaps for gene expression analysis – simple explanation with an example
In this post, you will learn how to interpret a heatmap for differential gene expression analysis. Find out why heatmaps are a great way of visualising gene expression data with this simple explanation. Let's dive in!Prefer to listen? Watch my Youtube video on...
Gene Set Enrichment Analysis (GSEA) – simply explained!
What is gene set enrichment analysis and how can you use it to summarise your differential gene expression analysis results?This post will give you a simple and practical explanation of Gene Set Enrichment Analysis, or GSEA for short. You will find out: What is Gene...
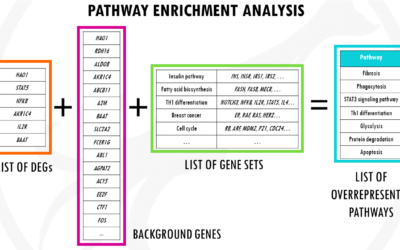
Pathway enrichment analysis for DGE – simply explained
An overview of pathway enrichment analysis and how you can use it for your differential gene expression analysis data. In this post, you will find pathway enrichment analysis explained in a simple way with examples. I will try to give you a simple and practical...
Multiple testing correction methods: FDR, q-values vs p-values
A simple explanation of what is multiple testing and how it can negatively affect your data. We will also cover some of the most common multiple testing correction methods.In this post I will try to give you a simple and practical explanation of multiple testing....
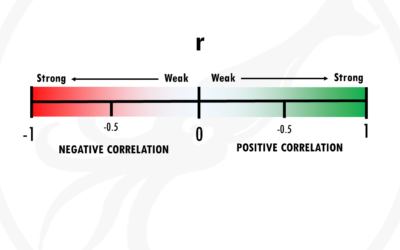
Correlation does not imply causation
Simple explanation of what is correlation, positive and negative correlation, and the correlation coefficient r.In this post I will try to give you a simple and practical explanation of correlation. Correlation is one of the most used statistical techniques. However,...
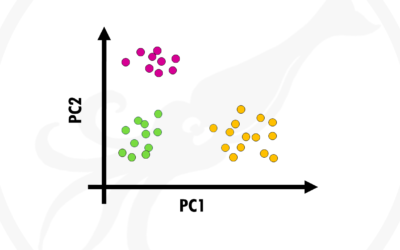
Principal Component Analysis (PCA) simply explained
In this post I will try to give you a simple and practical explanation on what is Principal Component Analysis and how to use it to visualise your biological data. Principal Component Analysis, or PCA, is a widely used technique to visualise multidimensional datasets....
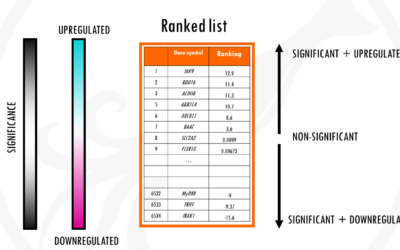
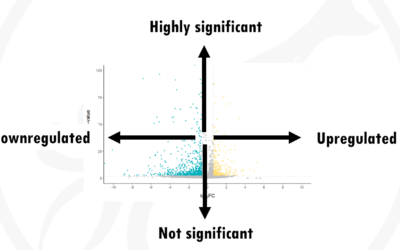
How to interpret a volcano plot
What is a volcano plot?Imagine you are carrying out an RNAseq experiment. You have a group of cells A and a group of cells B. Group B was treated with a drug. Now you want to see what effect the drug has in gene expression. Does the drug cause some genes to be...
How to choose log2FC thresholds for DGE analysis
Setting thresholds for differential gene expression (DGE) analysis is crucial and depends on several factors. In essence, for a list of genes, we are trying to define what counts as biologically meaningful versus just statistically significant. The question is... How...
Comparing multiple groups: Kruskal-Wallis test in R
When working with biological data, we often want to compare measurements across multiple groups. However, these measurements aren't always normally distributed. In such cases, non-parametric methods like the Kruskal-Wallis test and Dunn’s post-hoc test are ideal...
Understanding Seurat objects – simply explained!
Understanding the structure of Seurat objects version 5 - step-by-step simple explanation!If you've worked with single-cell RNAseq data, you've probably heard about Seurat. In this blogpost, we'll cover the the Seurat object structure,in particular the new Seurat...
SCTransform – simple and intuitive explanation
SCTransform (Single-Cell Transform) is a normalization method primarily used in scRNA-seq data analysis. It was developed to address limitations in standard normalization approaches when dealing with single-cell data. You can check how to apply SCTransform on your...
Why do genes with the highest logFC not have the lowest p-value?
That's a really good and very common question in differential gene expression analysis! It feels intuitive that the larger the difference in expression (log fold change, or logFC), the more significant it should be (i.e., the smaller the p-value), but that’s not...
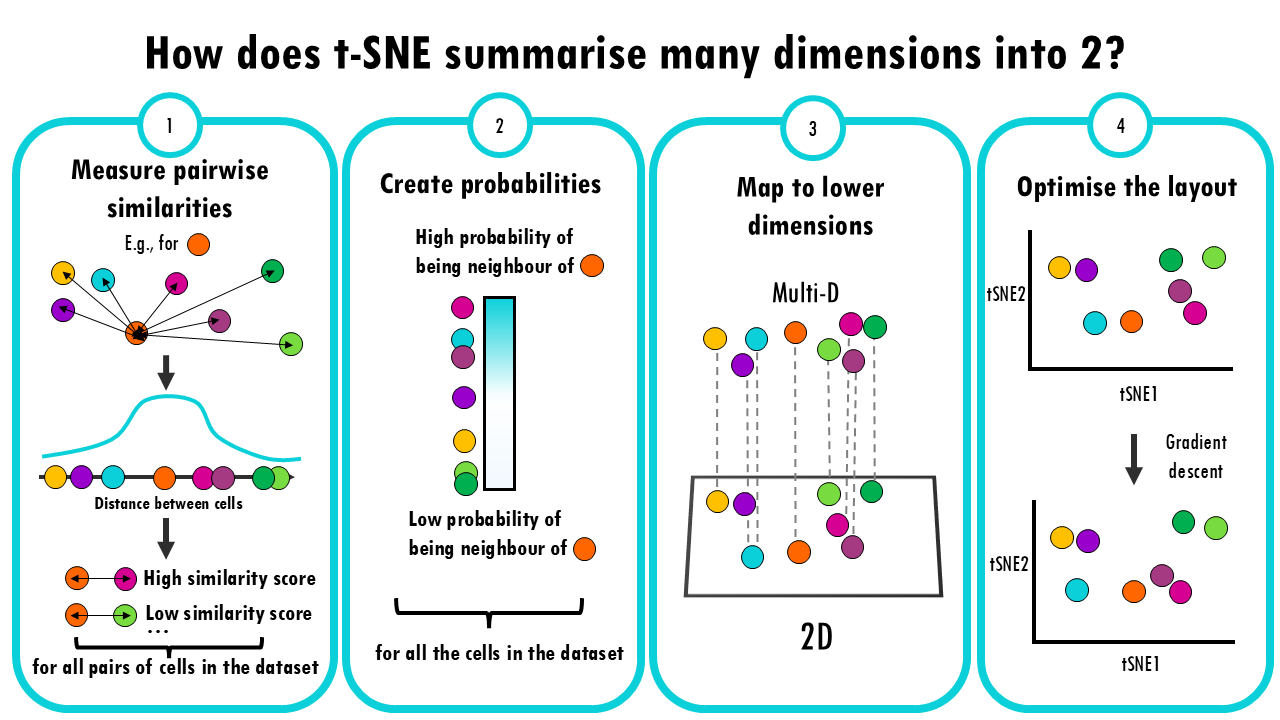
PCA vs UMAP vs t-SNE
Understanding similarities and differences between dimensionality reduction algorithms: PCA, t-SNE and UMAPPCA, t-SNE, UMAP... you've probably heard about all these dimensionality reduction methods. In this series of blogposts, we'll cover the similarities and...