A step-by-step easy R tutorial to detect and remove doublets using scDblFinder in R
In this easy, step-by-step scDblFinder tutorial you will learn how to detect and remove doublets from scRNAseq data in R, using the R package (you guessed it!) scDblFinder .
First things first. What are doublets?
Doublets are artifacts that occur when two cells are captured together and sequenced as one. This means that in your matrix, for a given cell barcode, you are getting gene expression data from 2 different cells. These doublets can really confound your downstream analysis, so it is important to identify them during the preprocessing steps and remove them.
There are many tools to infer doublets from scRNAseq data. You can check this doublet detection tools benchmarking paper here.
One of the tools I have found works well and is easy to implement is scDblFinder. You can read more about the tool and methods here and here.
In this tutorial, we will learn how to:
- run scDblFinder on a sce or seurat object
- compare some basic single-cell stats like number of genes, % mitochondrial genes…, between doublets and singlets
- mark and remove doublets from your dataset
We will use the R packages:
- tidyverse
- Seurat
- scDblFinder
If you are more of a video-based learner, you can check out my Youtube video on how to use scDblFinder in R. Otherwise, just keep reading! And remember you can just copy the code by clicking on the top right corner of the code snippets in this post.
Let’s dive in!
Check out my Youtube video to follow this scDblFinder in R tutorial with me!
Before you start…
In this tutorial, we will learn how to run scDblFinder in R. Remember you can refer to the paper and the GitHub page to read on the methods and options:
https://github.com/plger/scDblFinder
As always, you can find the code I am using in this tutorial at biotatsquid’s github page.
For this tutorial I’m going to be using a Seurat object that’s already preprocessed, meaning I did some preliminary QC, including filtering low quality cells and lowly expressed genes. If you are interested in a full scRNAseq preprocessing tutorial, check out this other post!
You can install scDblFinder from Bioconductor:
BiocManager::install("scDblFinder")Set up your environment
It’s good practice to clear up your environment before you load in any data. This is the general structure I like to keep in all my scripts:
# ---------------------- #
# scRNAseq doublet detection tutorial
# ---------------------- #
# Setting up environment ===================================================
# Clean environment
rm(list = ls(all.names = TRUE)) # will clear all objects including hidden objects
gc() # free up memory and report the memory usage
options(max.print = .Machine$integer.max, scipen = 999, stringsAsFactors = F, dplyr.summarise.inform = F) # avoid truncated output in R console and scientific notation
We also need to load the necessary libraries.
# Loading relevant libraries
library(tidyverse) # includes ggplot2, for data visualisation. dplyr, for data manipulation.
library(Seurat)
library(scDblFinder)Set the relevant paths
I have a project path (a folder called “scRNAseq_tbr”), and inside it I have a folder called data, which will be my input path. I create an out_path called results, where output all my plots and tables from the analysis. I’m going to create an additional subfolder called doublet_detection. That’s where all the doublet analysis results will be saved to. If this analysis is part of a bigger project you might want to set up a different folder system.
# Relevant paths ======================================================================
project_path <- "~/Biostatsquid/Projects/scRNAseq_tbr"
in_path <- file.path(project_path, 'data')
out_path <- file.path(project_path, 'results') # for all scRNAseq preprocessing results
doublet_folder <- file.path(out_path, 'Doublet_detection') # subfolder for Doublet detection results
#dir.create(doublet_folder, recursive = TRUE, showWarnings = FALSE)
Squidtip
Make sure those folders exist! Otherwise R will throw and error when you try to read data from or save data to them!
You can create the directories from within R using dir.create.
Explore the dataset
We’ll start by loading the data, we will load the Seurat object. As I mentioned it’s an intermediary Seurat object which I already processed: I filtered out low quality cells and genes expressed in very few cells.
# Inputs ======================================================================
# Read in your Seurat object
seu <- readRDS(file.path(in_path, "seu_filt.rds"))
head(seu@meta.data)
table(seu$SampleID)Let’s have a look at the metadata – as you can see there’s 2 samples in this Seurat object, one called Kidney and one called Lung. This is important because most doublet detection tools work on a per-sample basis, meaning that if your Seurat object has more than 1 sample, you need to split it per sample, find doublets in each subset, and then merge the datasets again.
> table(seu$SampleID)
Kidney Lung
4974 5036
The code in this tutorial will work if you have 1 or more than 1 sample.
Let’s start by detecting and analyzing doublets in single-cell RNA sequencing (scRNA-seq) data using the `scDblFinder` package in R.
Squidtip
The input to scDblFinder should not include empty droplets, and it might be necessary to remove cells with a very low coverage (e.g. <200 or 500 reads) to avoid errors. That is why it is important to preprocess and filter your cellxgene matrix to remove empty droplets before running scDblFinder.
1. Run scDblFinder
scDblFinder takes a SingleCellExperiment object as input. If you are not familiar with this type of object, you can read more about it here. If you already have your scRNAseq data in this format, you can just skip the next step and go directly to running scDblFinder, otherwise, we need to transform the Seurat object to SingleCellExperiment class.
# Doublet detection =================================================================
## scDblFinder -------------------------------
# Run scDbltFinder
sce <- as.SingleCellExperiment(seu)Next, we will run scDblFinder. We can do this really easily by just using the function scDblFinder(). The `samples` parameter is used to specify the metadata column with the sample IDs, if you have more than one sample. You can also finetune the expected multiplet rate with the parameter `dbr` and `dbr.sd`. Check out scDblFinder’s github page to find out more or run “?scDblFinder”!
sce <- scDblFinder(sce, samples = "SampleID") #dbr = multiplet_rate)
table(sce$scDblFinder.class)
sce@colData@listData %>% as.data.frame() %>% head()> table(sce$scDblFinder.class)
singlet doublet
9094 916 2. Summary of doublet detection results
Let’s have a look at how many doublets and singlets we found. The labels will be stored in the metadata column called “scDblFinder.class”.
If we take a look at the metadata, we see scDblFinder outputs 5 columns, which are prefixed by “scDblFinder”. Let’s extract these columns into a separate dataframe. We have:
- scDblFinder.sample: the sample IDs
- scDblFinder.class: the labels (singlet or doublet)
- scDblFinder.score: the final doublet score (the higher the more likely that the cell is a doublet). I won’t go into the details of their method, but have a look at their documentation because you can finetune the threshold to call a doublet based on the expected number of doublets. I went with the default this time.
- scDblFinder.weighted: score based on the proportion of the KNN that are doublets, weighted by their distance (useful for isolated cells)
- scDblFinder.cxds_score: score based on an estimate of the difficultly of detecting artificial doublets in the cell’s neighborhood, a variant of the
cxdsscore from the scds.
# Explore results and add to seurat object
meta_scdblfinder <- sce@colData@listData %>% as.data.frame() %>%
dplyr::select(starts_with('scDblFinder')) # 'scDblFinder.class')
head(meta_scdblfinder)3. Add scDblFinder results to Seurat object
If you were working with a SingleCellExperiment class object, you can skip this step.
Now, if you started off with a Seurat object like I did, we need to add the doublet labels to the original Seurat object. So we first extract the metadata, then we make sure that the dataframe’s rows are the same as the ones in the Seurat object – add the cellIDs as rownames.
And now we can add the metadata with the function AddMetaData(). I’m just going to add the scDblFinderclass, but you can add the other columns as well by removing this part. Feel free to customise the name of the column.
# Explore results and add to seurat object
meta_scdblfinder <- sce@colData@listData %>% as.data.frame() %>%
dplyr::select(starts_with('scDblFinder')) # 'scDblFinder.class')
head(meta_scdblfinder)
rownames(meta_scdblfinder) <- sce@colData@rownames
head(meta_scdblfinder)
seu <- AddMetaData(object = seu, metadata = meta_scdblfinder %>% dplyr::select('scDblFinder.class'))> head(meta_scdblfinder)
scDblFinder.sample scDblFinder.class scDblFinder.score scDblFinder.weighted
Kidney_TCAATTGGTCGAAGTAACTTTAGG-1 Kidney singlet 0.011418721 0.39223541
Kidney_AACCTGCTCACGTTCAACTTTAGG-1 Kidney singlet 0.165400475 0.55228446
Kidney_CGCATTTCAAAGAACAACTTTAGG-1 Kidney singlet 0.002969444 0.24925287
Kidney_TGTGAGGTCATGGATAACTTTAGG-1 Kidney singlet 0.002201870 0.02223416
Kidney_CCTATGATCTGTCAGCACTTTAGG-1 Kidney singlet 0.001550194 0.15758762
Kidney_TGTACCGGTACGCACCACTTTAGG-1 Kidney doublet 0.998503804 0.97904009
scDblFinder.cxds_score
Kidney_TCAATTGGTCGAAGTAACTTTAGG-1 0.0980932600048948777349266947567230090498924255371093750000000000000000000000000000
Kidney_AACCTGCTCACGTTCAACTTTAGG-1 0.0000000000000000000000000000000000000000000000000000000000000000000000000000450728
Kidney_CGCATTTCAAAGAACAACTTTAGG-1 0.0082297947742535765280891268957930151373147964477539062500000000000000000000000000
Kidney_TGTGAGGTCATGGATAACTTTAGG-1 0.0001279967934597507964438606320456415232911240309476852416992187500000000000000000
Kidney_CCTATGATCTGTCAGCACTTTAGG-1 0.0008653346509565277688141859968595781538169831037521362304687500000000000000000000
Kidney_TGTACCGGTACGCACCACTTTAGG-1 0.0822368214796967700319640925954445265233516693115234375000000000000000000000000000
Just to make sure, let’s check again the number of singlets and doublets.
We can also now remove the temporary variables `meta_scdblfinder` and `sce` from the environment to clear up a bit of memory.
And now you can visualise some stats!
head(seu@meta.data)
table(seu$scDblFinder.class)
rm(list = c('meta_scdblfinder', 'sce'))> table(seu$scDblFinder.class)
singlet doublet
9094 916 4. scDblFinder QC stats and summary table
For example, le’ts plots violin plots of quality control metrics (number of RNA features, RNA counts, mitochondrial percentage, ribosomal percentage, hemoglobin percentage) split by `scDblFinder` class and grouped by `SampleID`. As you can see doublets tend to have a higher number of genes and read counts, but we don’t expect big differences in the mitochondrial percentage or ribosomal percentage.
# Doublet stats
# Check how doublets singlets differ in QC measures per sample.
VlnPlot(seu, group.by = 'SampleID', split.by = "scDblFinder.class",
features = c("nFeature_RNA", "nCount_RNA", "percent_mt", "percent_ribo", "percent_hb"),
ncol = 3, pt.size = 0) + theme(legend.position = 'right')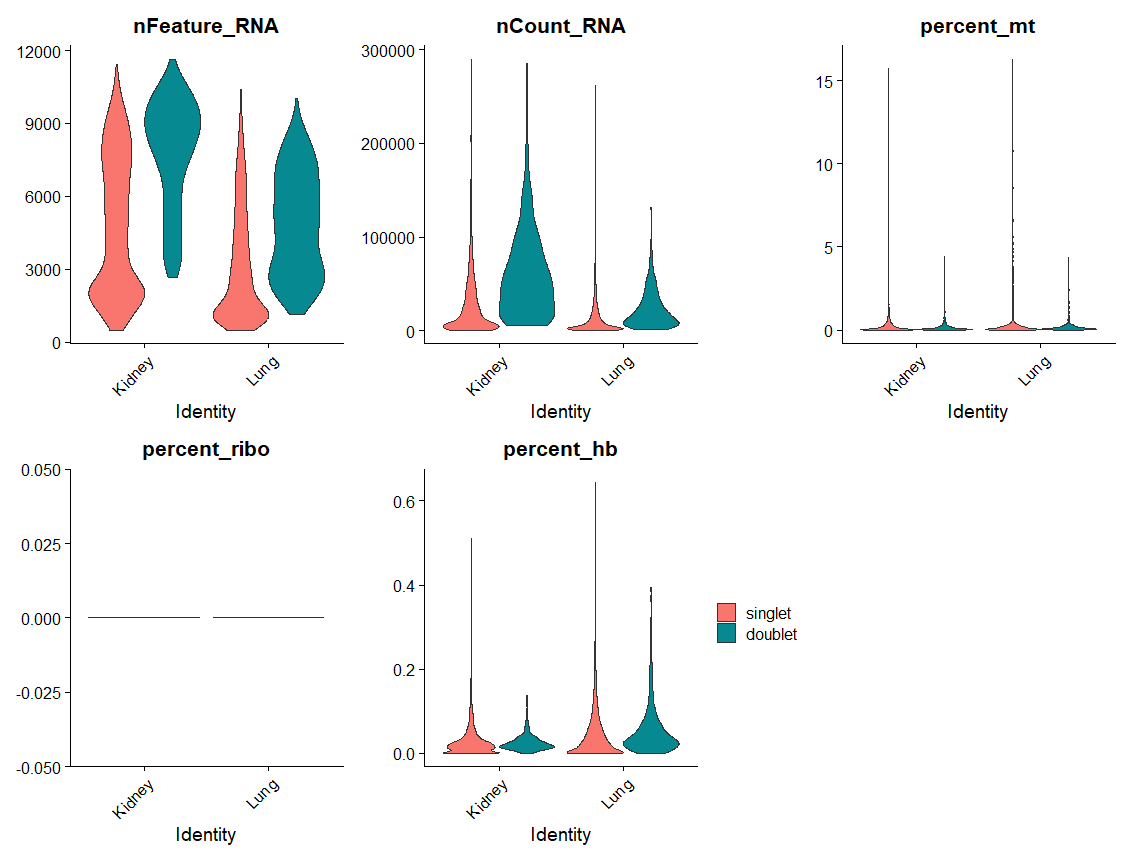
We can also create a nice summary table by grouping the metadata by `SampleID` and `scDblFinder.class` and calculating the total count of cells in each group and the percentage of doublets and singlets for each sample.
And of course we can save this summary table to a text file.
doublets_summary <- seu@meta.data %>%
group_by(SampleID, scDblFinder.class) %>%
summarise(total_count = n(),.groups = 'drop') %>% as.data.frame() %>% ungroup() %>%
group_by(SampleID) %>%
mutate(countT = sum(total_count)) %>%
group_by(scDblFinder.class, .add = TRUE) %>%
mutate(percent = paste0(round(100 * total_count/countT, 2),'%')) %>%
dplyr::select(-countT)
doublets_summary
write.table(doublets_summary, file = file.path(doublet_folder, paste0('scDblFinder_doublets_summary.txt')), quote = FALSE, row.names = FALSE, sep = '\t')> doublets_summary
# A tibble: 4 × 4
# Groups: SampleID, scDblFinder.class
[4]
SampleID scDblFinder.class total_count percent
<chr> <fct> <int> <chr>
1 Kidney singlet 4526 90.99%
2 Kidney doublet 448 9.01%
3 Lung singlet 4568 90.71%
4 Lung doublet 468 9.29% 5. Remove doublets
You can remove cells marked as “doublet” very easily with the function subset().
# Removing doublets ============================================================
# If all is ok subset and remove doublets
seu_dblt <- subset(scDblFinder.class == 'singlet')And that’s it! We have successfully detected and removed doublets with scDblFinder in R!
sessionInfo()
Check my sessionInfo() here in case you have trouble reproducing my steps:
> sessionInfo()
R version 4.3.2 (2023-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 11 x64 (build 22631)
Matrix products: default
locale:
[1] LC_COLLATE=English_Ireland.utf8 LC_CTYPE=English_Ireland.utf8 LC_MONETARY=English_Ireland.utf8 LC_NUMERIC=C
[5] LC_TIME=English_Ireland.utf8
time zone: Australia/Sydney
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices datasets utils methods base
other attached packages:
[1] DoubletFinder_2.0.4 scDblFinder_1.12.0 SingleCellExperiment_1.24.0 SummarizedExperiment_1.32.0 Biobase_2.62.0
[6] GenomicRanges_1.54.1 GenomeInfoDb_1.38.5 IRanges_2.36.0 S4Vectors_0.40.2 BiocGenerics_0.48.1
[11] MatrixGenerics_1.14.0 matrixStats_1.2.0 Seurat_5.0.1 SeuratObject_5.0.1 sp_2.1-2
[16] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2
[21] readr_2.1.5 tidyr_1.3.1 tibble_3.2.1 ggplot2_3.4.4 tidyverse_2.0.0 And that is the end of this tutorial!
This is a very simple workflow for detecting, summarizing, and visualizing doublets in scRNA-seq data using Seurat and scDblFinder in R.
When detecting doublets in scRNAseq data, you often want to test out several tools and even finetune parameters for each tool. Another tool I’ve tried that has given me great results and is relatively easy to use is DoubletFinder. You can find a tutorial here. In this other blogpost, I compare both tools and remove only high-confidence doublets (doublets that were found by both tools).
You can read more abut DoubletFinder here.
Before you go, you might want to check:
- scDblFinder publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9204188/
- scDblFinder documentation: https://bioconductor.org/packages/devel/bioc/vignettes/SingleCellExperiment/inst/doc/intro.html
- scDblFinder github: https://github.com/plger/scDblFinder
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and… see you in the next one!
Squids don't care much for coffee,
but Laura loves a hot cup in the morning!
If you like my content, you might consider buying me a coffee.
You can also leave a comment or a 'like' in my posts or Youtube channel, knowing that they're helpful really motivates me to keep going:)
Cheers and have a 'squidtastic' day!


0 Comments