An overview of pathway enrichment analysis and how you can use it for your differential gene expression analysis data.
In this post, you will find pathway enrichment analysis explained in a simple way with examples.
I will try to give you a simple and practical explanation of PEA and how to use it to get the most out of your DGE data.
You will find out:
-
- What is Pathway Enrichment Analysis (PEA) and why is it a great way to use it for differential gene expression data
- What do you need to perform PEA
- The main idea behind all PEA algorithms
- How to interpret PEA results
- Common pitfalls when performing PEA and how to avoid them
If you are more of a video-based learner, you can check out my Youtube video, otherwise, just keep reading.
Let’s dive in!
Check out my Youtube video to learn the basics of Pathway Enrichment Analysis!
What is pathway enrichment analysis?
Differential gene expression (DGE) analysis is an essential step in RNAseq downstream analysis. The goal is to identify differentially expressed genes (DEGs) between two conditions.
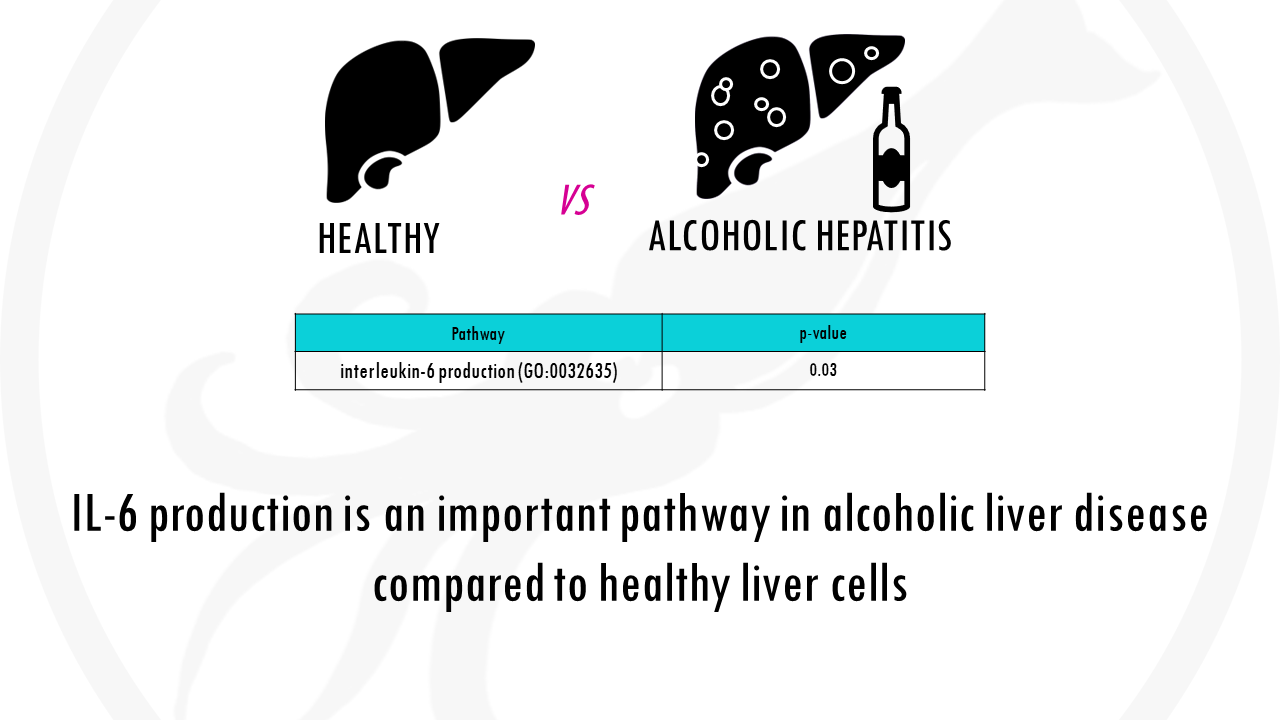
For example, you might be interested in studying the difference in gene expression between liver cells of healthy individuals and liver cells in individuals with alcoholic hepatitis.
But DGE analysis may return long lists of differentially expressed genes (e.g., in the order of hundreds or thousands).
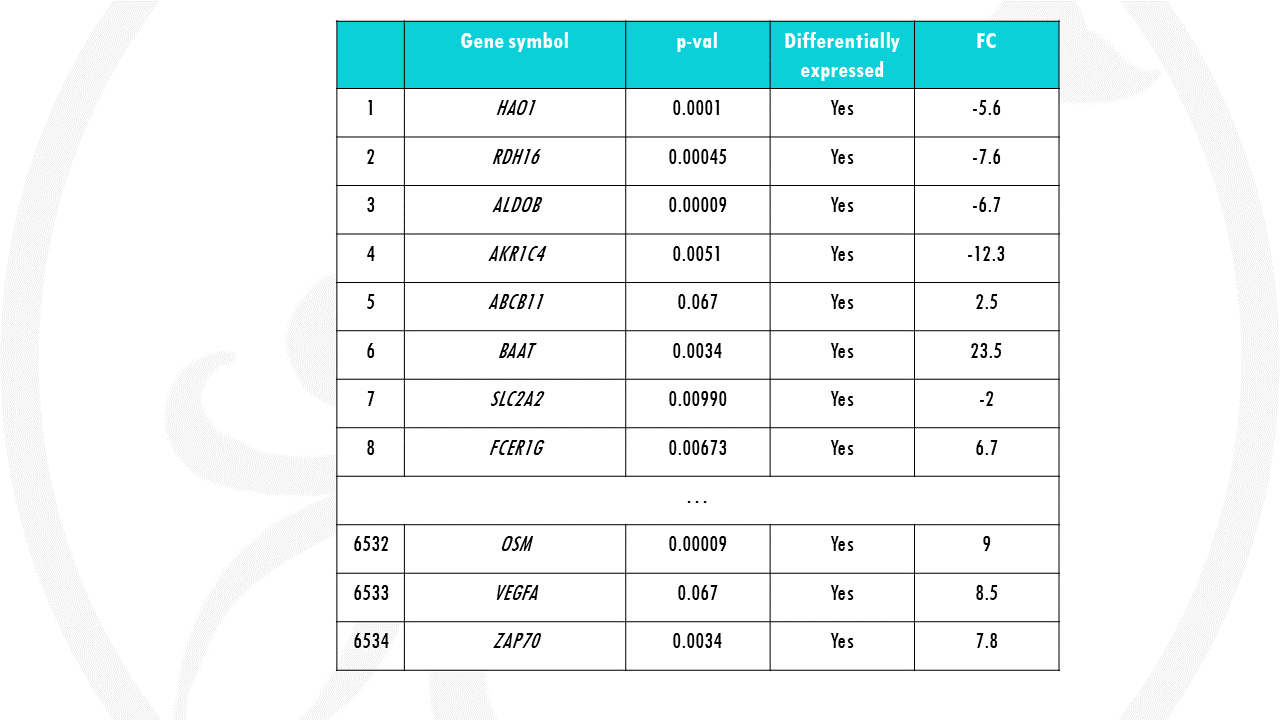
How do we even start interpreting this?
Is there a way to summarise this long list of genes and interpret hundreds of DGEs at once?
A common approach is pathway enrichment analysis (PEA).
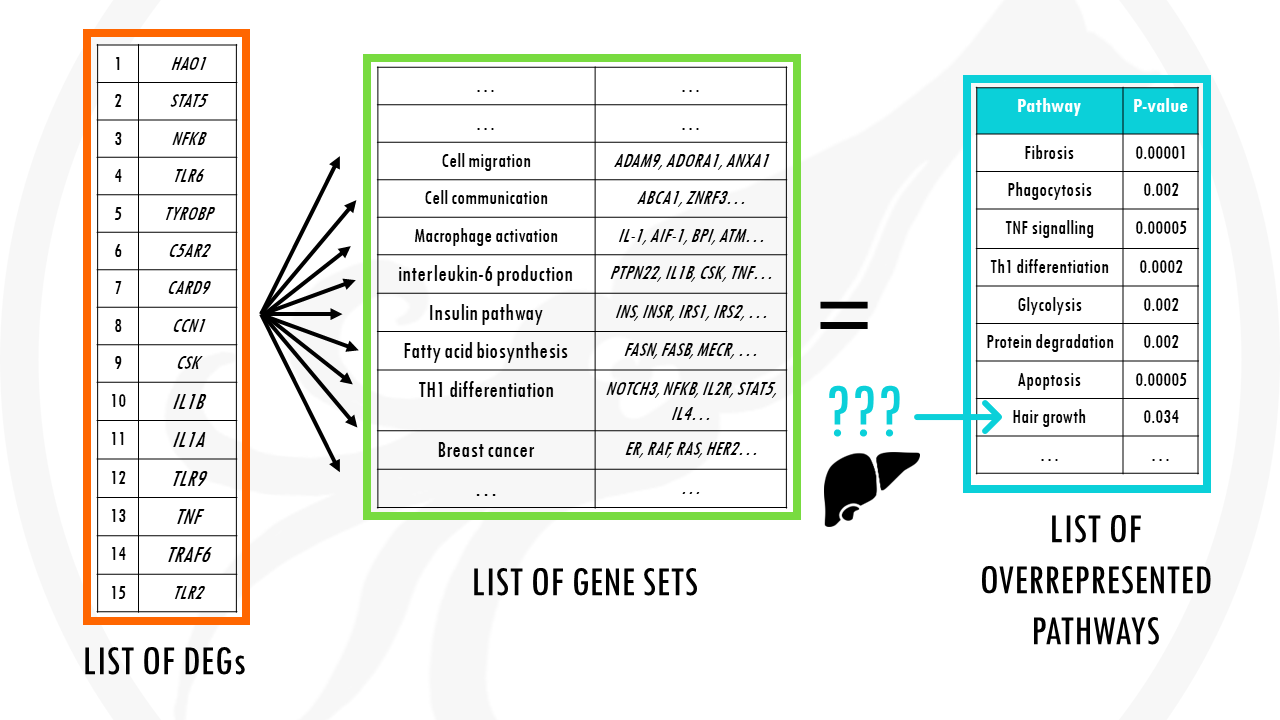
It basically summarises the long gene list to a shorter and more easily interpretable list of pathways.
So instead of having a list of 20.000 genes, you may get a list of 50 or 60 biological pathways. And of course, you can then check which genes are behind these pathways.

In one sentence,
Pathway enrichment analysis summarises the long gene list to a shorter and more easily interpretable list of pathways.
How does Pathway Enrichment Analysis work?
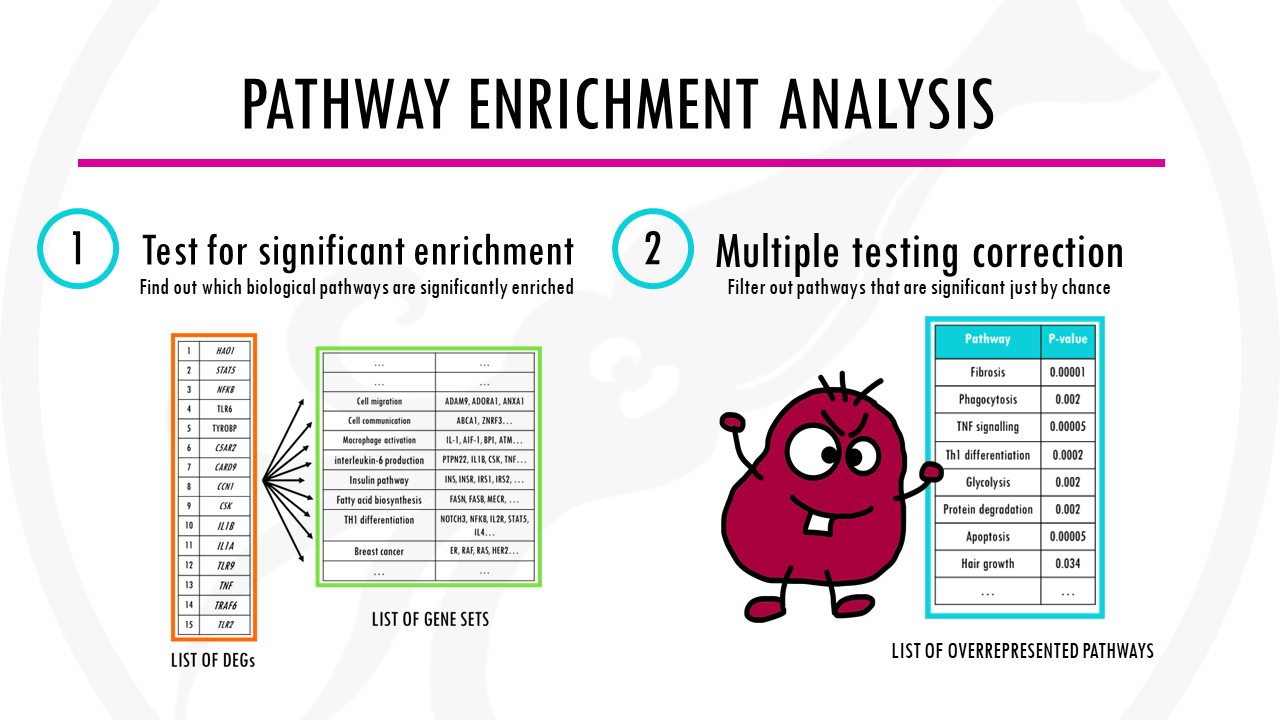
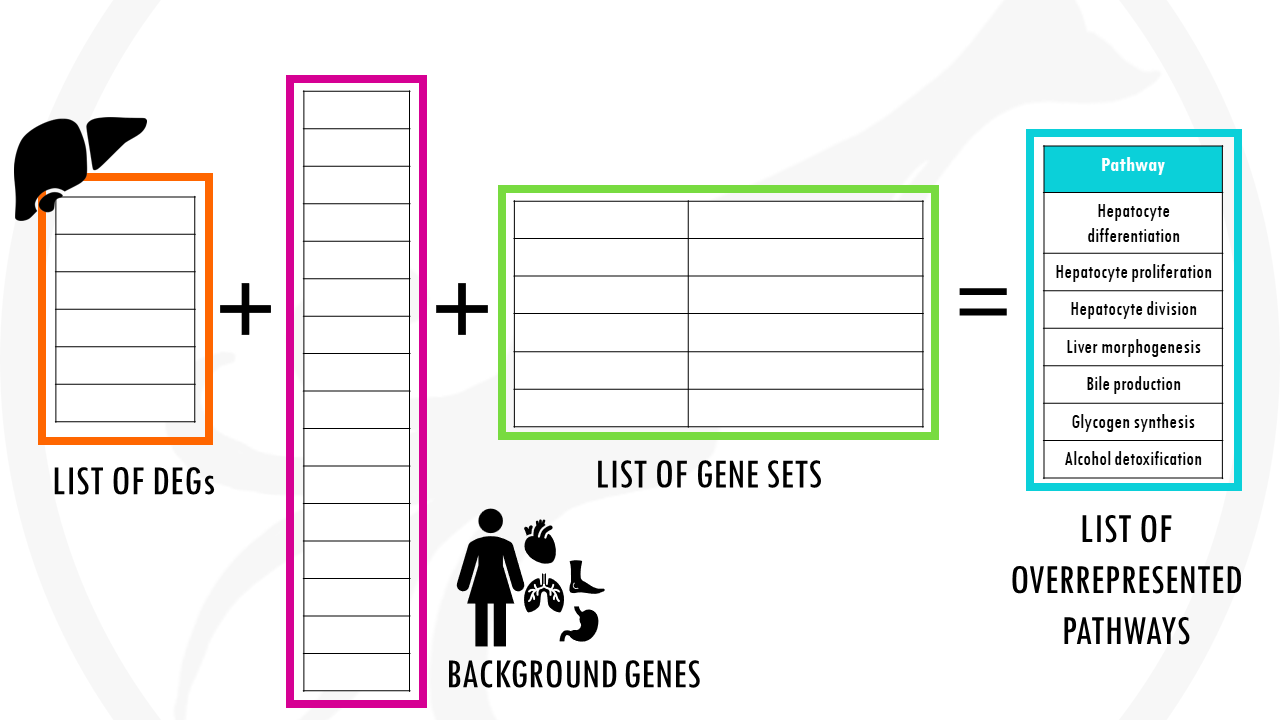
Pathway enrichment analysis needs 3 ingredients.
First, of course, your gene list of interest for example, a list of differentially expressed genes which you want to summarise.
Second, a list of background genes – for example, all of the genes in the human genome.
Finally, it will take a lists of gene sets. Gene sets are basically groups of related genes. Of course, for the algorithm to know if your list has a lot of genes related to breast cancer, or apoptosis, or cellular respiration, you need to tell it which genes are actually involved in breast cancer, apoptosis, and cellular respiration.
You will find out a bit more about each component later on.
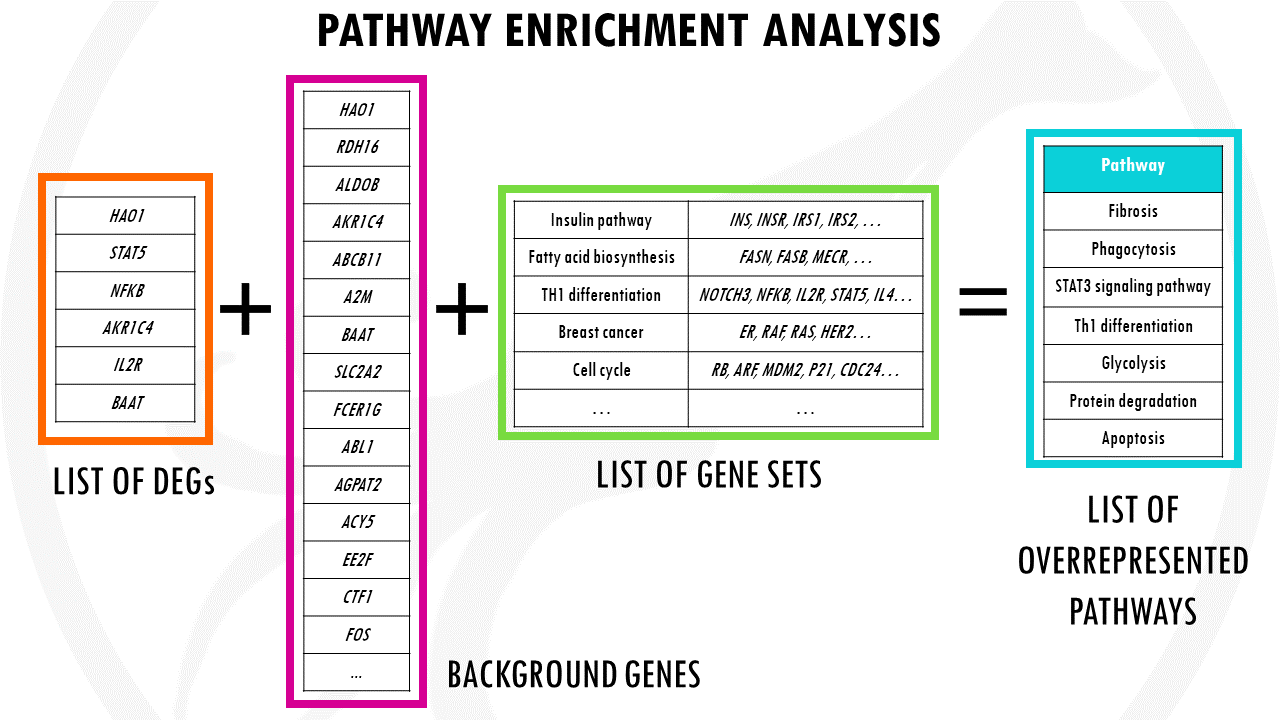
PEA essentially compares your gene list to the background list to check if there are certain pathways overrepresented.
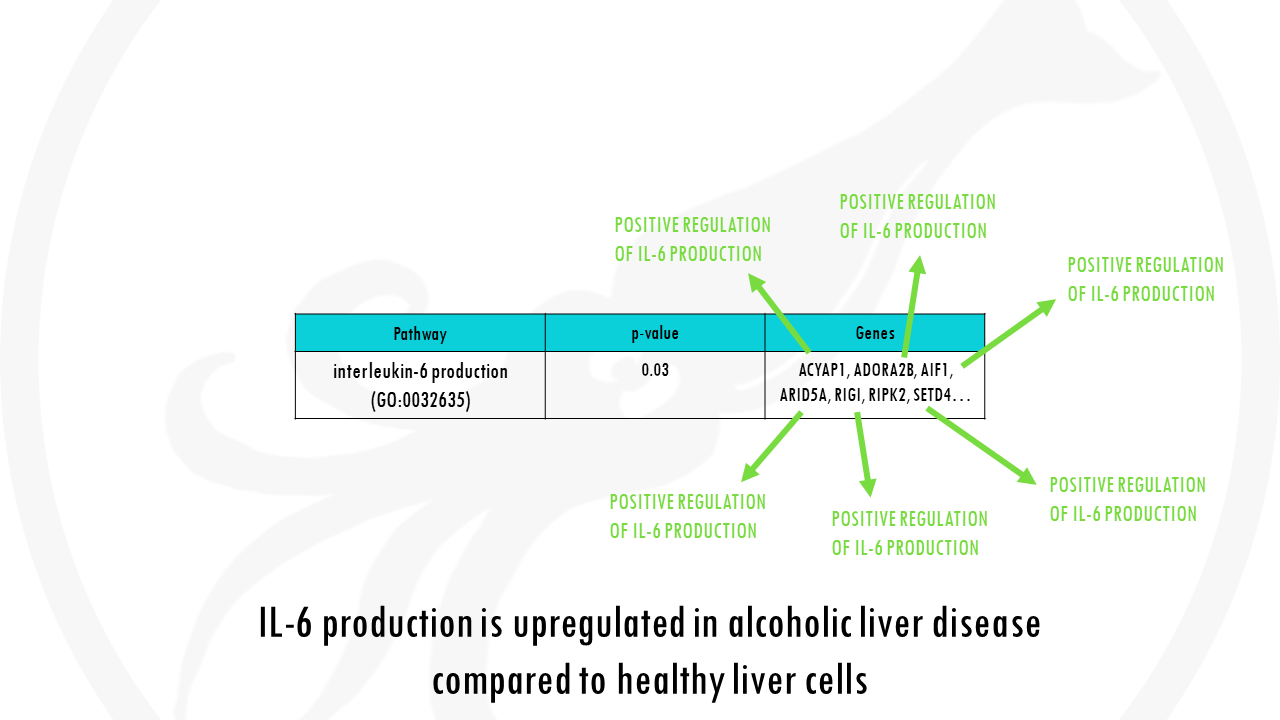
Let’s go back to our example. Alcoholic liver disease is usually involved in inflammatory processes, which often involve pro-inflammatory cytokines like IL-6.
So, is there an association between our genes differentially expressed in alcoholic liver disease vs healthy cells and interleukin-6 production?
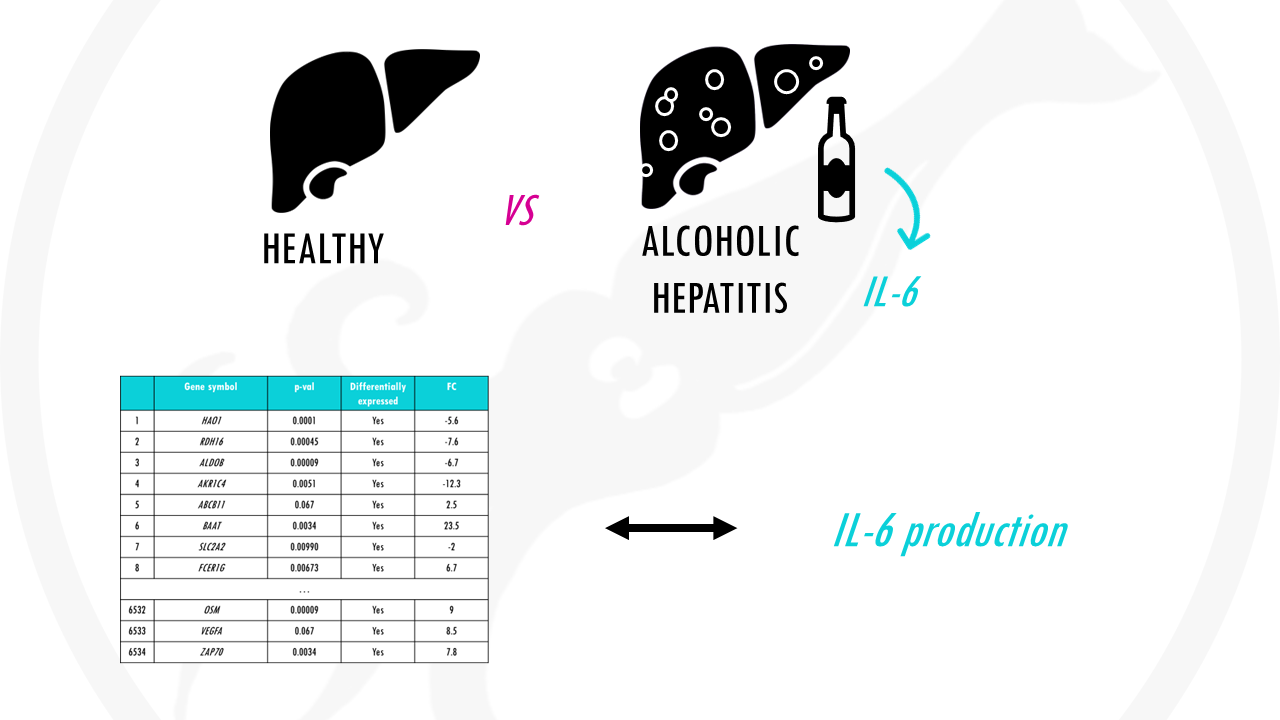
In other words, is our list of differentially expressed genes enriched with genes involved in IL-6 synthesis pathway?
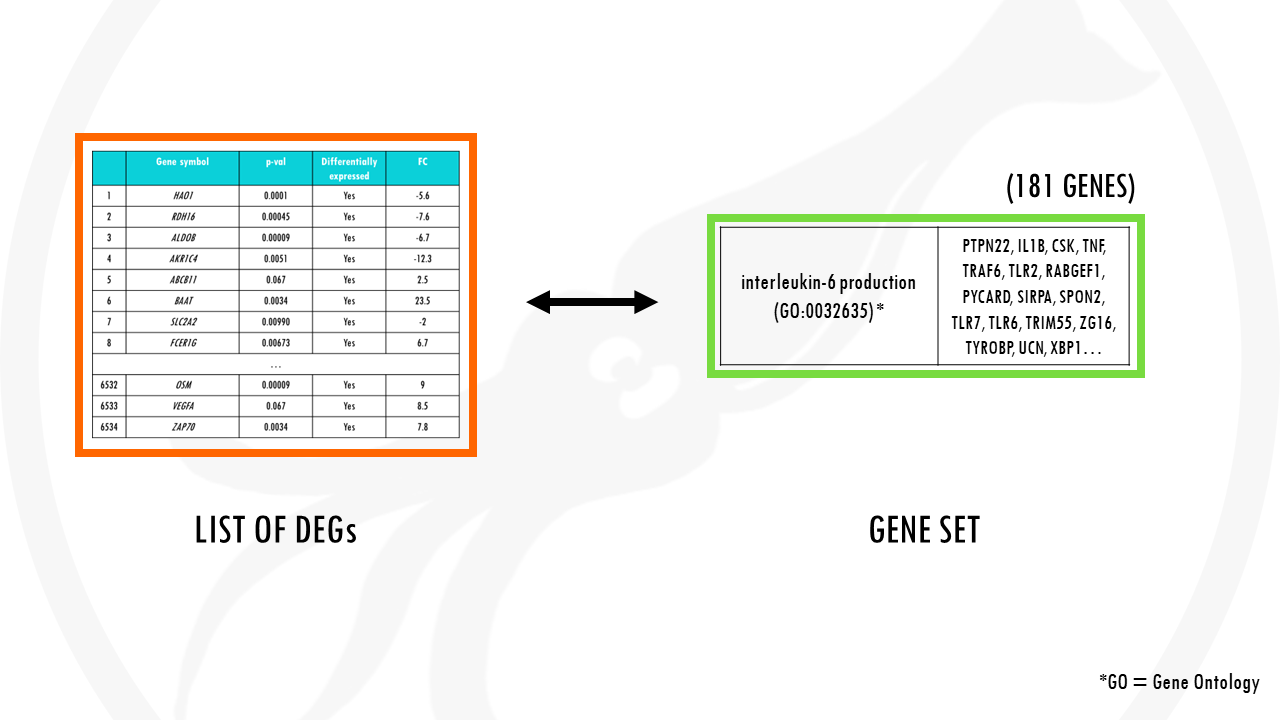
To answer this question, we can build a contingency table. This will help us determine whether the fraction of genes of interest in the pathway is higher compared to the fraction of genes outside the pathway (so, background set).
You can have a look at the table below.
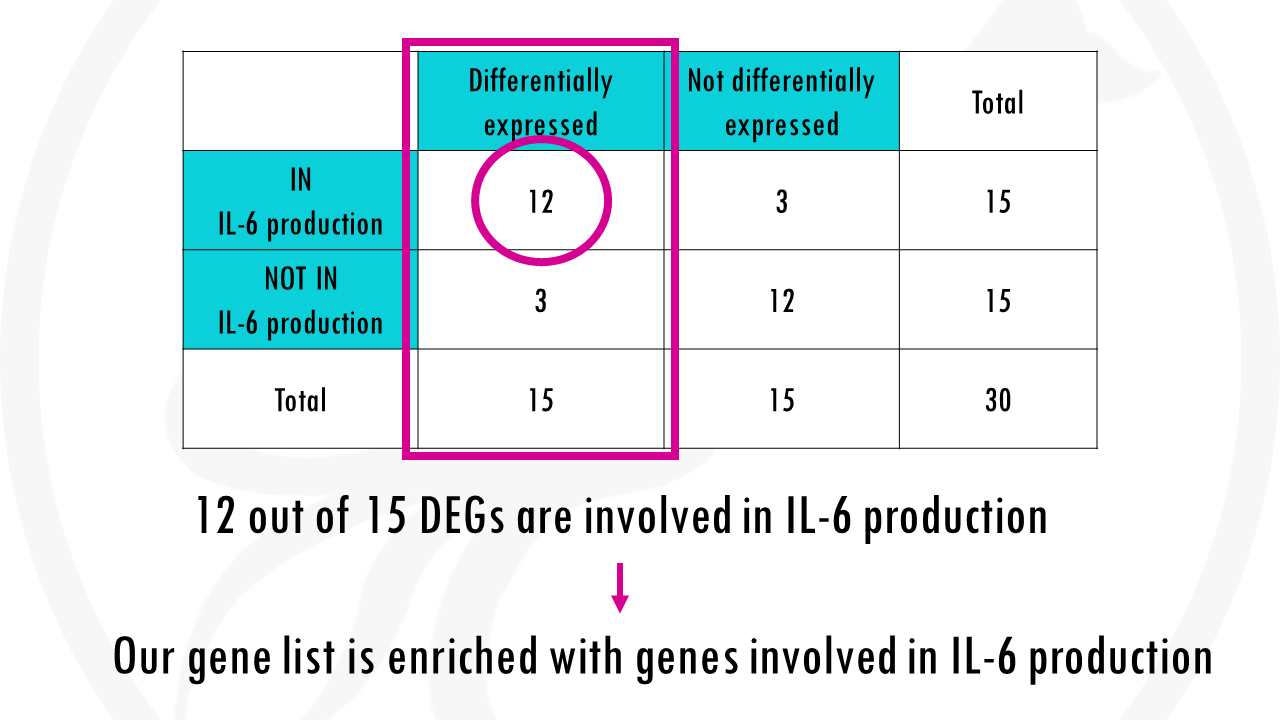
We have a column for differentially expressed and a column not differentially expressed genes, and then two rows, for genes that are annotated as being involved in IL-6 production and genes that are not involved in IL6 production.
To simplify things a lot, we will just look at 30 genes. 15 differentially expressed genes were identified and of those, 12 genes were associated with the GO term interleukin-6 production.
We find that 12 out of our 15 differentially expressed genes are involved in interleukin-6 production. We could quite confidently say that our gene list is enriched with genes involved in IL-6 production.
Ok, but what if they were 9 out of 15 genes?
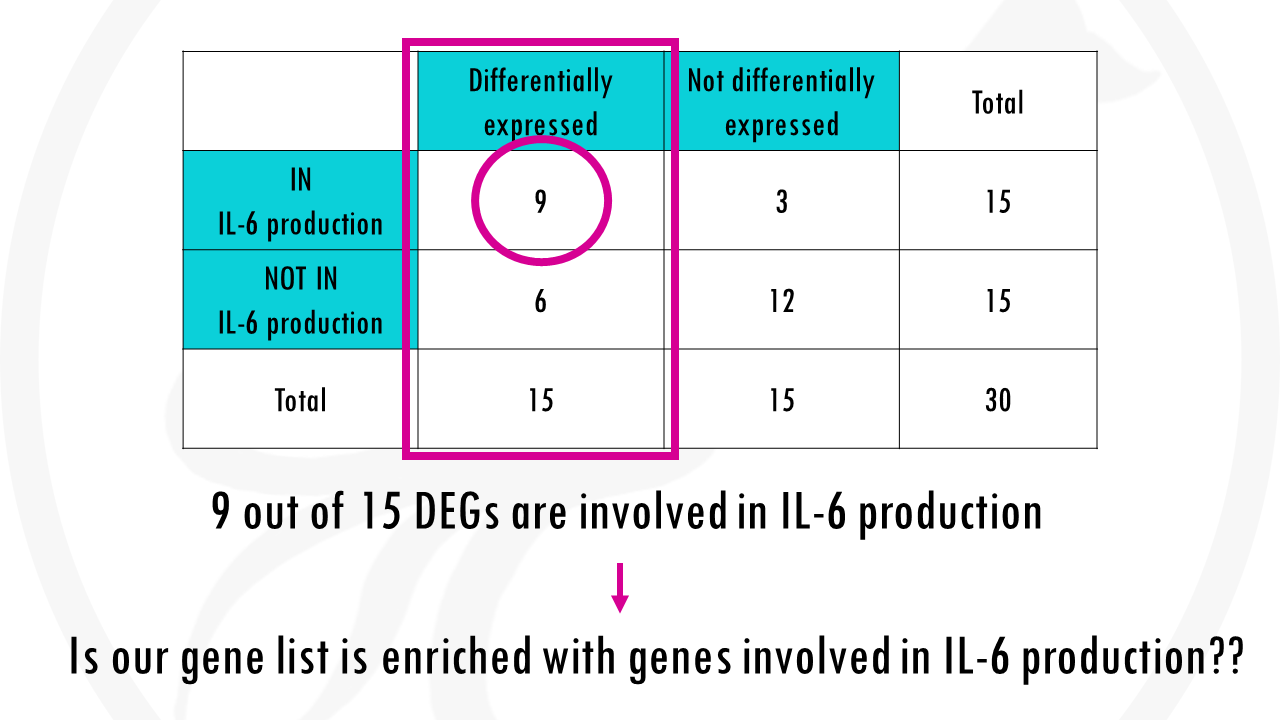
Obviously, we need an objective statistical test to determine what is enriched and what is not.
There are many methods out there, but the one that is commonly used in pathway enrichment analysis is Fisher’s exact test.
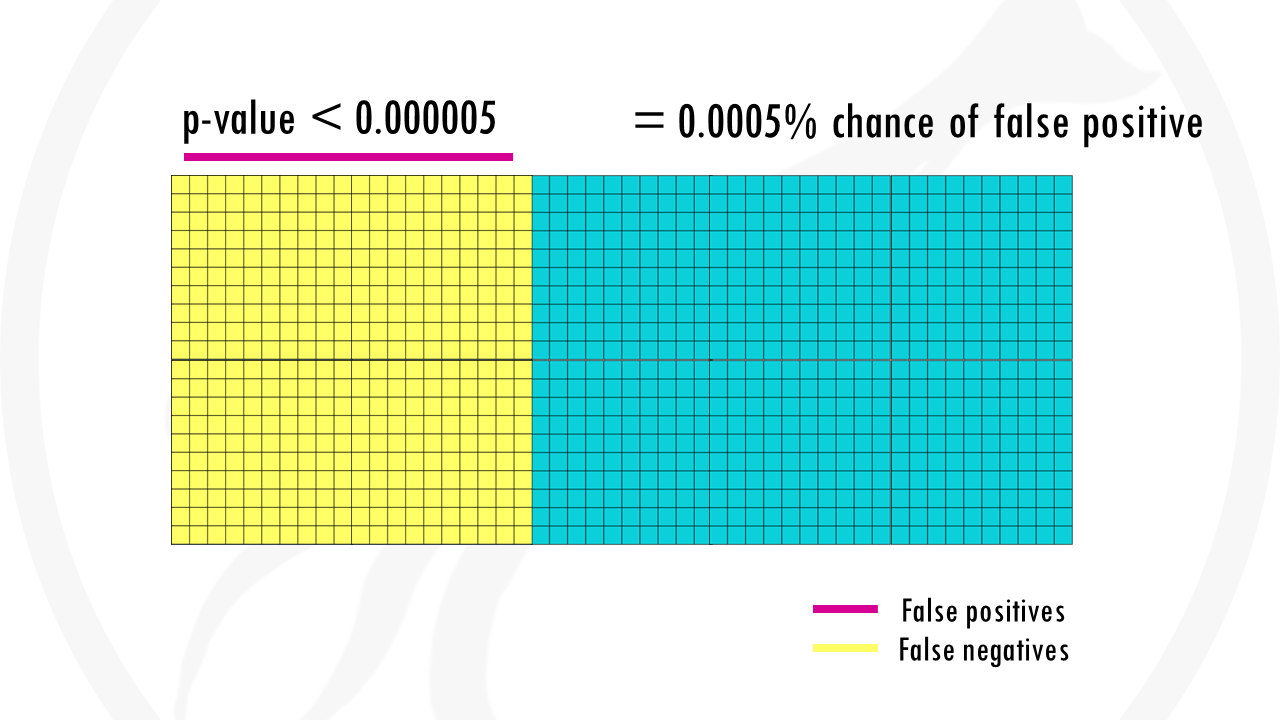
Like with most statistical tests, you will obtain a p-value.
If you p-value is really low, you can safely say that your list is overrepresented with genes involved in IL-6 production, in other words, IL-6 production is an important pathway in alcoholic liver disease compared to healthy liver cells.
Squidtip
Careful! A low p-value does not mean that pathway is upregulated! You would have to check the genes that are actually overrepresented in your list, and see if they are positive or negative regulators of that pathway.
So this is what pathway enrichment is all about.
You summarise a long list of genes to a shorter list of pathways, with their p-values.
Obviously, it does it not with one, but you with thousands of pathways.
And this bring us to a BIG problem.
The big problem is called multiple testing.
Basically, because we are repeatedly testing a lot of pathways, some pathways will get apparently significant p-values just by chance.
So we might get results that are a bit unexpected… or that just don’t make any sense.
Thank goodness there is a solution for this.
We just need a multiple-testing correction method. The most commonly used method is the Benjamini-Hochberg (BH) correction.
If you are not familiar with multiple testing or would like to know more, I suggest this other post. I also have a Youtube video that explains it with a bit more detail.
In any case, enrichment tools will both test for significant enrichment, and correct for multiple testing.
So, in summary
Pathway enrichment analysis takes your gene list of interest and compares it to a list of background genes to check if there are certain pathways that are overrepresented.
So it checks the fraction of your genes annotated to a specific Gene Ontology (GO) term. Then it checks the proportion of genes in the whole genome (your background set) that are annotated to that GO Term.
Then, it gives you a p-value which tells you what is the probability that that pathway is actually overrepresented in your gene list and it wasn’t just coincidence.

To be exact, the p value of a pathway is the probability or chance of seeing at least x number of genes out of the total n genes in the list annotated to a particular gene set term – for example, Th1 differentiation – , given the proportion of genes in the whole genome that are annotated to that gene set term.
The closer the p-value is to zero, the more significant the particular GO term associated with the group of genes is (i.e. the less likely the observed annotation of the particular GO term to a group of genes occurs by chance).
Gene sets
Let’s talk a bit more about your gene sets.
There are many databases of gene sets out there.
Some of the most widely used are Gene Ontology GO, KEGG or Reactome.
- GO basically focuses on biological processes
- KEGG is more focused on metabolic pathways
- Reactome is a curated database of human molecular pathways…

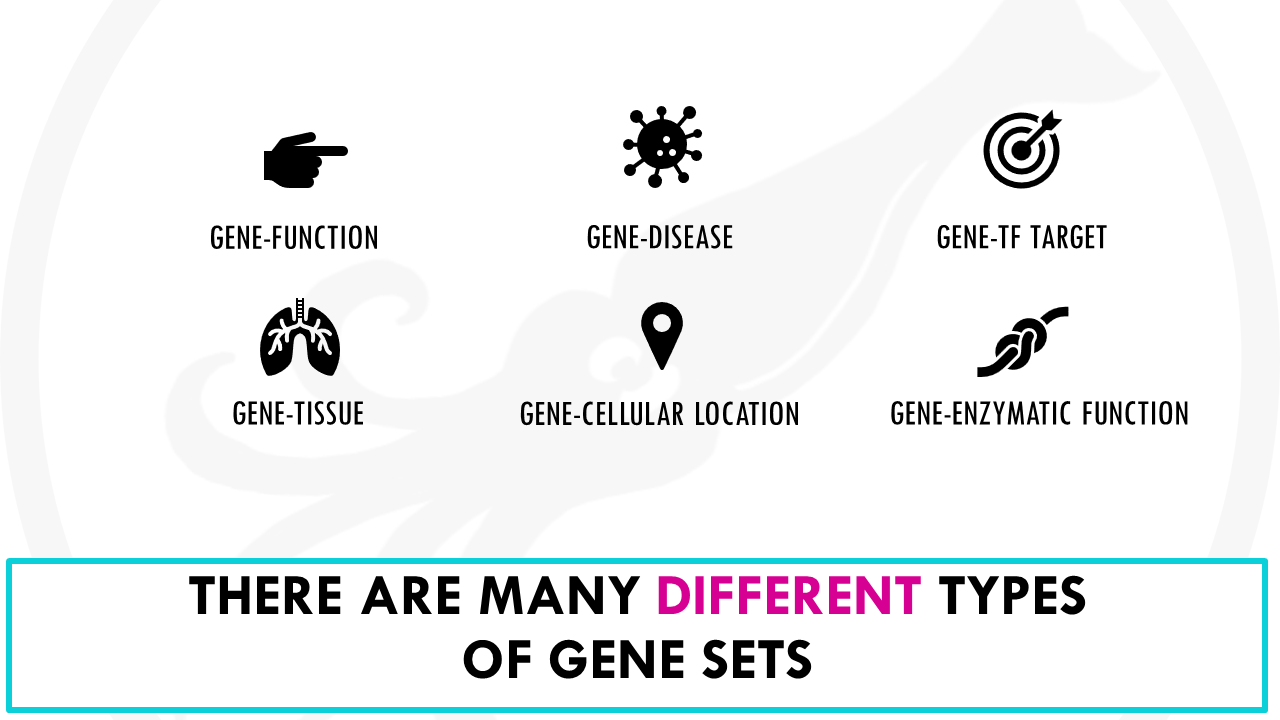
But gene sets are not restricted to functions.
There are gene sets for diseases, which gives you groups of genes associated to different diseases, tissues, which gives you groups of genes expressed in specific tissues, transcription factor targets, which tells you which genes are the target of different transcription factors…
Background genes
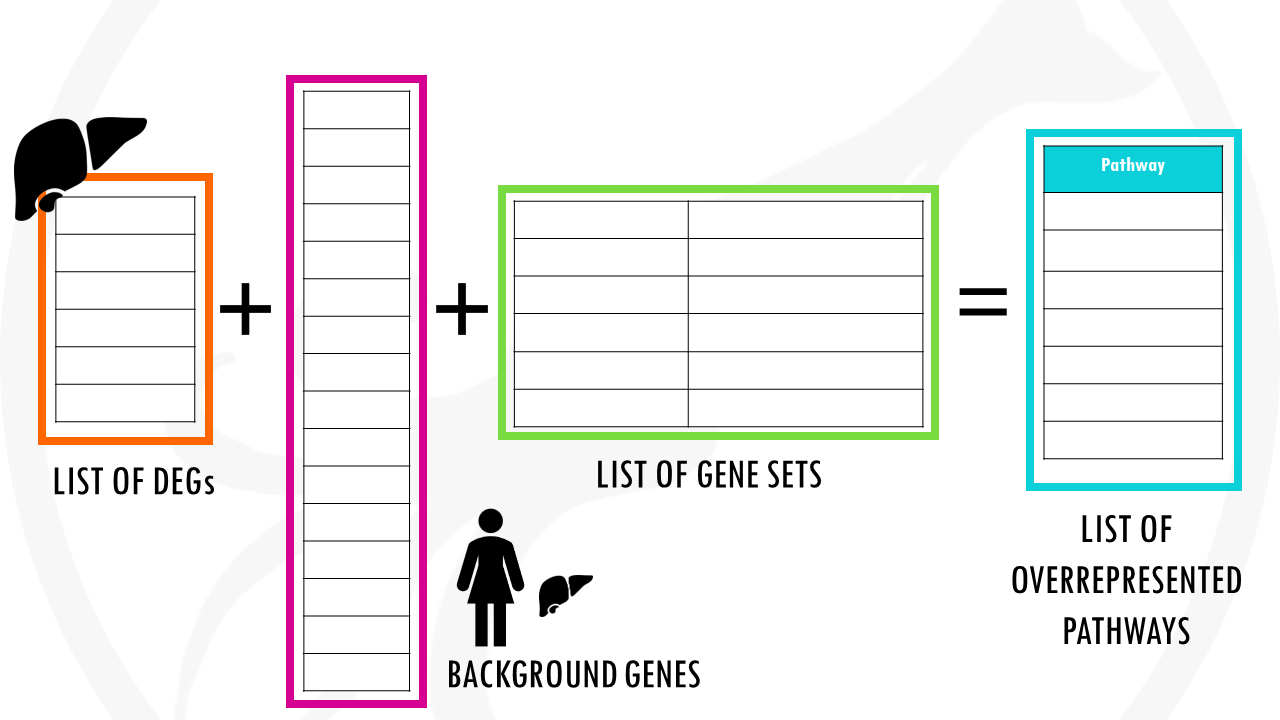
It is essential to choose well your background genes to your experiment.
Let’s go back to our example of liver cells.
If we use all the genes in the human genome as a background, and we perform enrichment analysis, it will probably tell us that our list of differentially expressed genes is enriched in liver function pathways.
I mean it is true, but it is not very helpful.
Instead, we might want to use as a background list, genes expressed in liver tissue and remove those that are never expressed in liver, for example, heart-specific genes.
That way, it will tell us which specific functions of liver cells are overrepresented in our analysis.
So we need to choose a custom background gene set that can be measured in the experiment.
If we’re dealing with liver cells, then exclude genes that are specifically expressed in other tissues.
Phosphoproteomics experiments measure only proteins with one or more phosphorylation sites. So our background gene set should only include genes encoding phosphoproteins. Otherwise, pathway enrichment analysis would reveal inflated P values for general processes such as kinase signalling and protein phosphorylation. You get the point.
Gene list
Finally, our gene list.
Results from gene expression analysis often look like this. Some genes are downregulated, some are upregulated, some don’t change. Some changes are significant, and some are not significant at all.
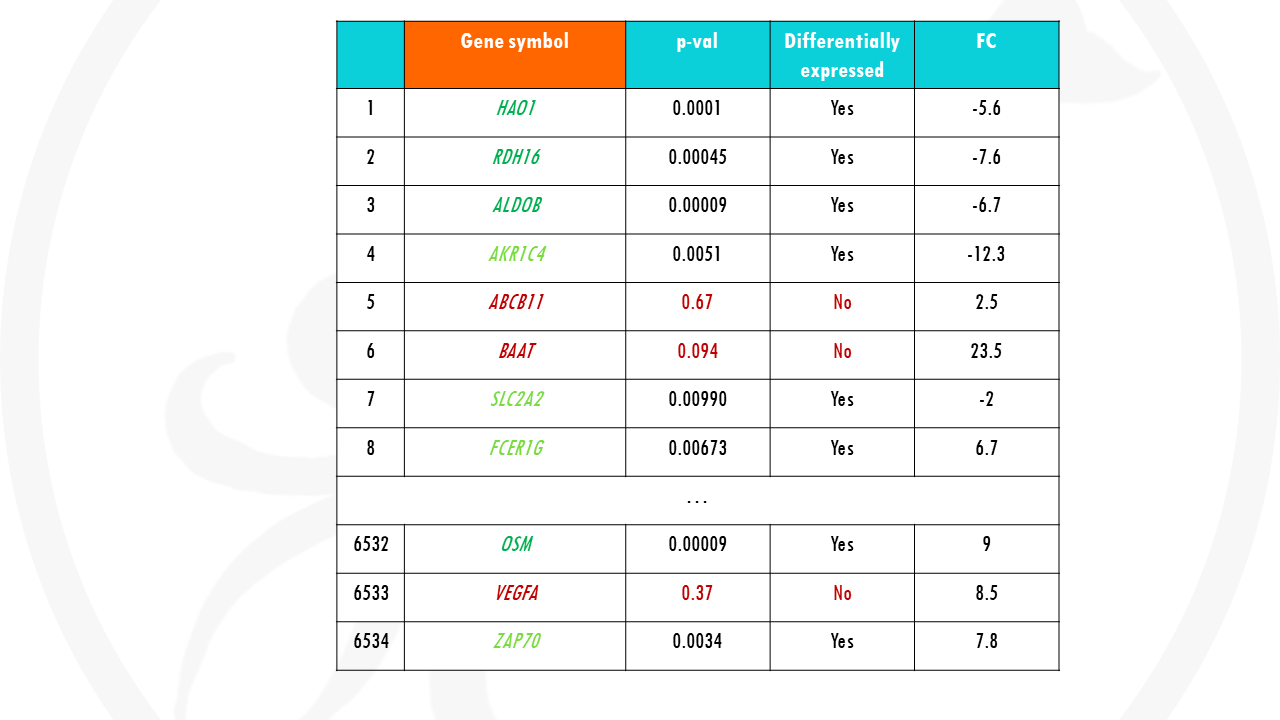
If you just use this list for pathway enrichment analysis, it will not take into account all that information. It will also match genes that are not even differentially expressed.
So you need to first filter your results, by significance and fold change, to keep only differentially expressed genes. The genes of interest.
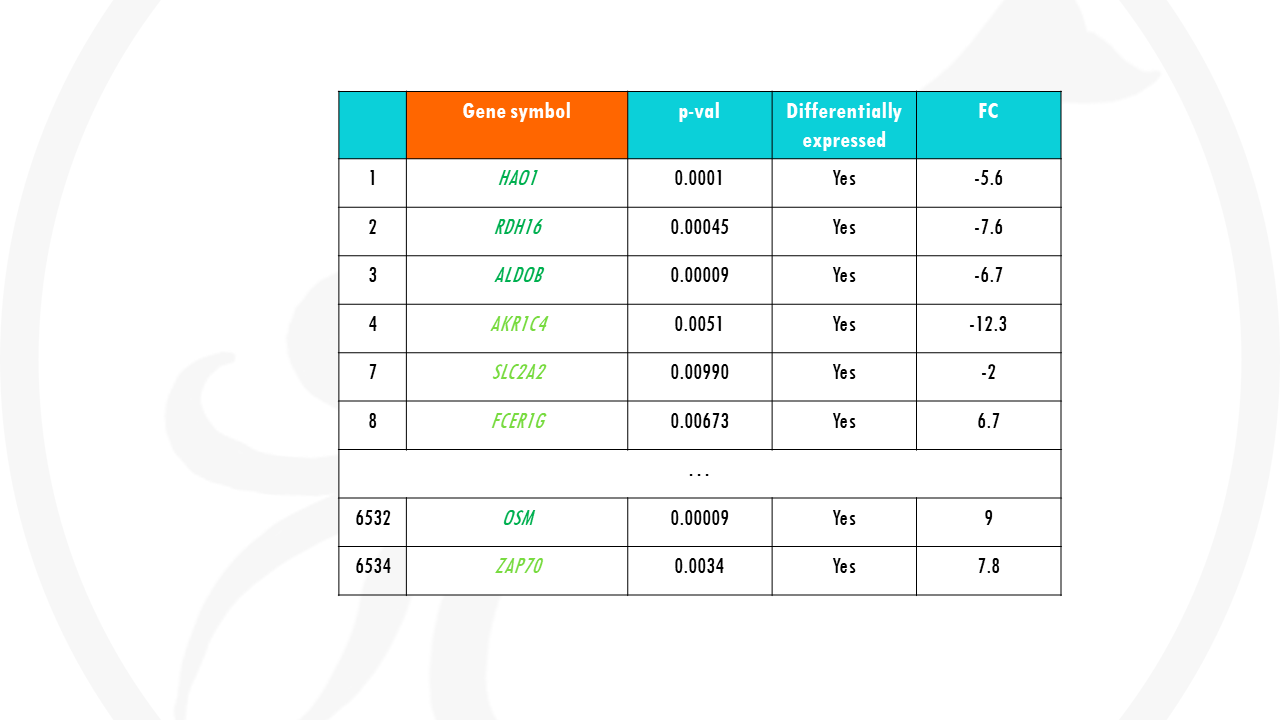
Of course, the results can change a lot depending on the cut-offs you set to say what is a DEG and what is not.
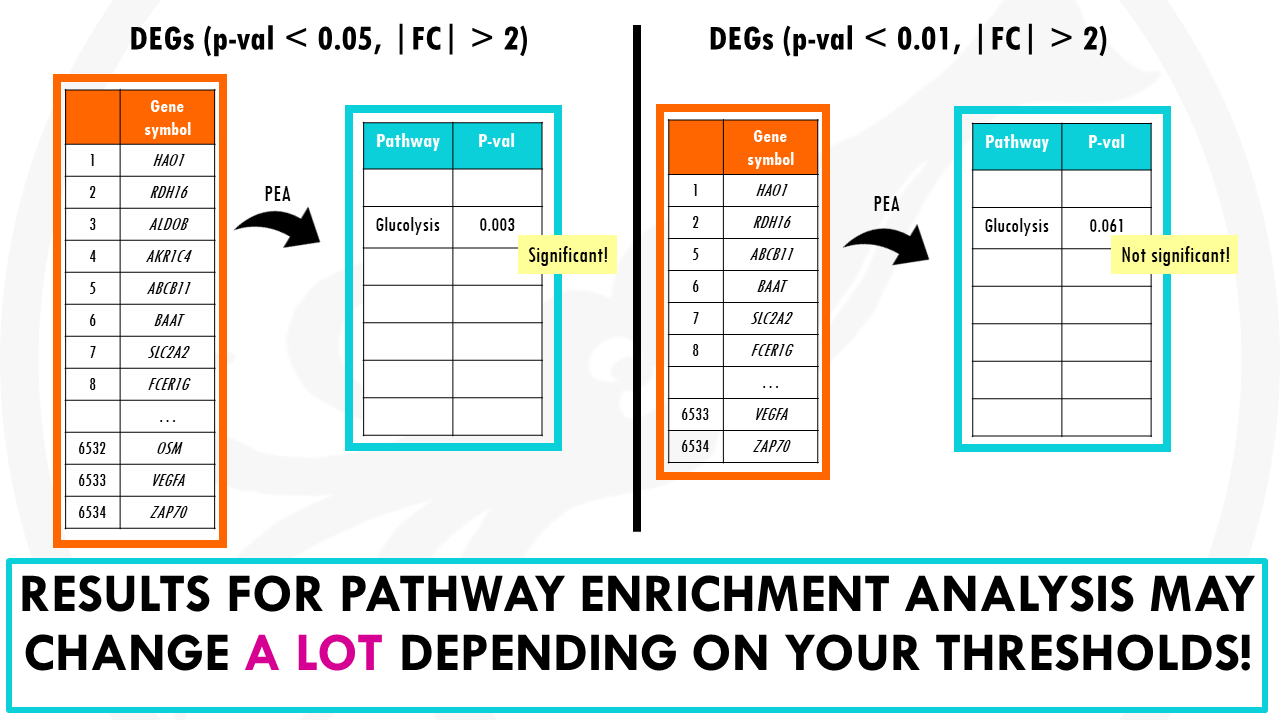
Is there a more objective or unbiased way of doing this?
For example, by taking into account the significance and strength of the changes, which is information we have anyway?
Of course there is!
The method is called Gene Set Enrichment Analysis and it a great pathway enrichment analysis method which helps us solve this problem. Find more about it in this other post.
Squidtastic!
And that is the end of this tutorial!
In this post, I explained the differences between log2FC and p-value, and why in differential gene expression analysis we don't always get both high log2FC and low p-value. Hope you found it useful!
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and... see you in the next one!


thank you so much for making these posts! I find them really interesting and useful. They also have good illustrations! keep it up!
Hi Jose, thank you so much for taking the time to comment and for your lovely feedback. It’s great to know that my work is helping people:) Also really open and keen to hear suggestions on how I can improve or what you would like to see next! Cheers!
Hi!
Thank you so much for your post. Just one quick question: So, background genes will only take pathways that contain at least one of our data_genes… and if we proceed without filtering the gmt file (filtering those pathways without any of our ddata_genes) , it may lead to biased results right?
Thanks in advanced for your answer and cheers from Peru!.
Thank you so much for your contribution.
Exactly David, couldn’t have explained it better. If you’d like to know more, this article explains exactly that and recommends to definitely remove non-expressed genes from the background: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-015-0761-7
Thanks so much for your comment!!
Hi!
I tried to do it with and without filtering. The results are similar but there are differences. I only did this once but I got a higher NES score and a slightly more significant adjusted p value when using the filtered pathways. Also some people argue that background should include any genes that could have been positive. What are your thought on this?
Hi! Thanks for your comment!
It’s important to filter your genes for pathway enrichment analysis to the genes that could have been positive, as you well said. Check this thread for more: https://www.biostars.org/p/17628/
As part of my major project I was surfing web to understand enrichment analysis but ended up bewildered.Finally,your post and youtube video made things simple that too in an entertaining fashion.Thank you so much your post and effort to make bioinformatics simple and fun.I damn excited for your future posts and videos…All the very best for your future endeavours!!!
Thank you so much for your comment! I really enjoy making bioinformatic posts and videos so glad you like my content! Same to you:)