A short but simple explanation of UMAP- easily explained with an example!
PCA, t-SNE, UMAP… you’ve probably heard about all these dimensionality reduction methods. In this series of blogposts, we’ll cover the similarities and differences between them, easily explained!
In this post, you will find out what is UMAP and how to interpret it with an example.
You can check part 1: easy PCA here and part 2: easy t-SNE here.
So if you are ready… let’s dive in!
Click on the video to follow my easy UMAP explanation with an example on Youtube!
What is dimensionality reduction?
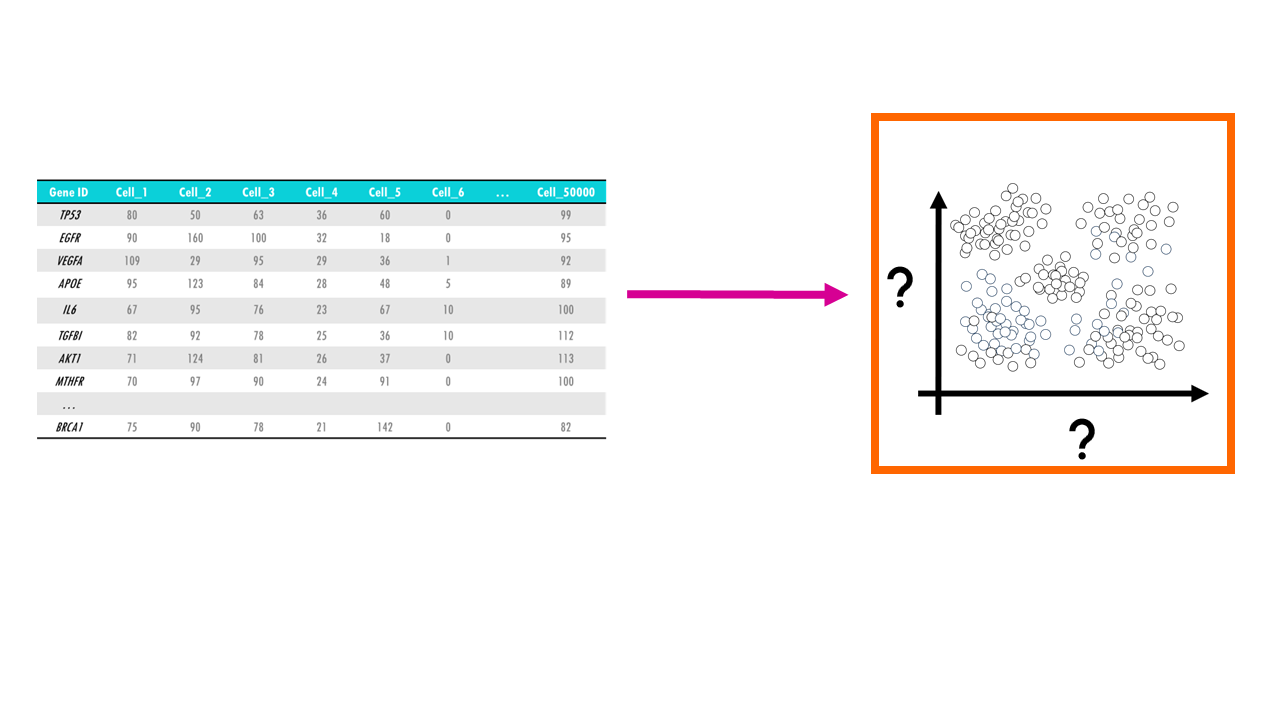
Let’s imagine you have a single-cell dataset with the gene expression of 10000 genes across 50.000 cells. For each cell, and for each gene, you have a gene expression value that tells you how much that gene was expressed in that cell.
Sometimes, we just want to visualise large datasets like this one in a plot. If each point in the plot is a cell, that would allow us to identify clusters of cells with similar expression levels of certain genes, we can identify cell types, or cells that are very different to the rest in their gene expression profile. The problem with such big datasets like this one, is that we cannot visualise all features, or genes, at once. A plot will have two axes, three at most. So we could pick two genes, and plot each cell according to their expression level of MCM3 and AKT1. We could then identify clusters of cells which have a high expression of both MCM3 and AKT1, and cells here have high expression of MCM3 but relatively low expression of AKT1, for example. However, this plot leaves out the expression level of the other 9998 genes in our dataset.
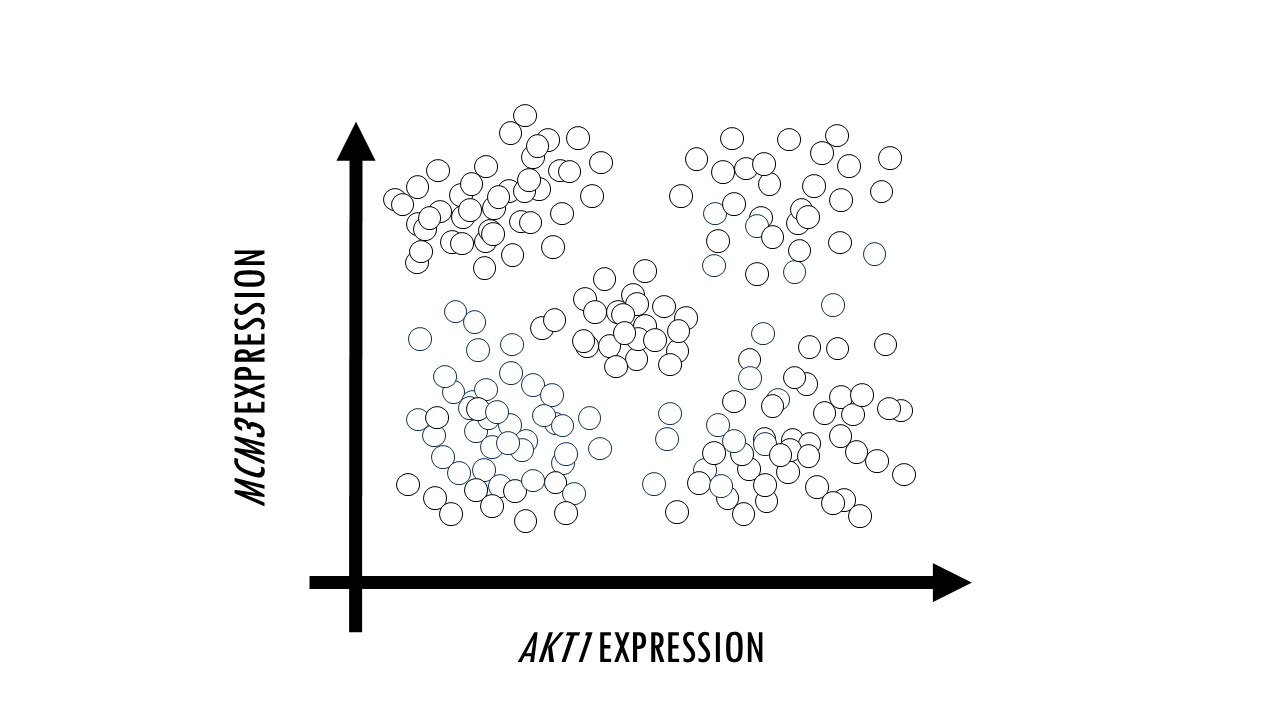
In other words, we need a way to convert a multidimensional dataset into two variables we can plot. This process is called dimensionality reduction and through it we basically reduce the number of input variables or features (in this case, genes) while retaining maximum information. Obviously, when we reduce the number of dimensions, we are going to lose information, but the idea is to preserve as much of the dataset’s structure and characteristics as possible.
To do this, we need a dimension reduction algorithm, which is basically a method that reduces the number of dimensions. As you can imagine, there are many dimension reduction algorithms out there, but we are going to talk about 3 of the most common ones: PCA, UMAP and t-SNE. We will go through the main ideas of each of them and the differences between them, so you can decide which one is most appropriate for your data.
Squidtip
Dimensionality reduction is a process that transforms data from high-dimensional space into a low-dimensional space while preserving as much of the dataset’s original information as possible.
UMAP (Uniform Manifold Approximation and Projection)
In a nutshell, UMAP is another dimensionality reduction technique, similar to t-SNE, but it’s often faster and can better preserve global structure while still focusing on local relationships.
Just like t-SNE, UMAP tries to take high-dimensional data and map it into a lower-dimensional space (usually 2D or 3D) so we can visualise it. It does this by preserving both local and global structures, meaning it tries to keep clusters of cells close together but also respects the overall organization of the data.
Squidtip
UMAP is a non-linear dimensionality reduction algorithm particularly well-suited for visualizing high-dimensional data by modelling each high-dimensional object by a two- or three-dimensional points (UMAP projections) in such a way that similar objects are modelled by nearby points and dissimilar objects are modelled by distant points.
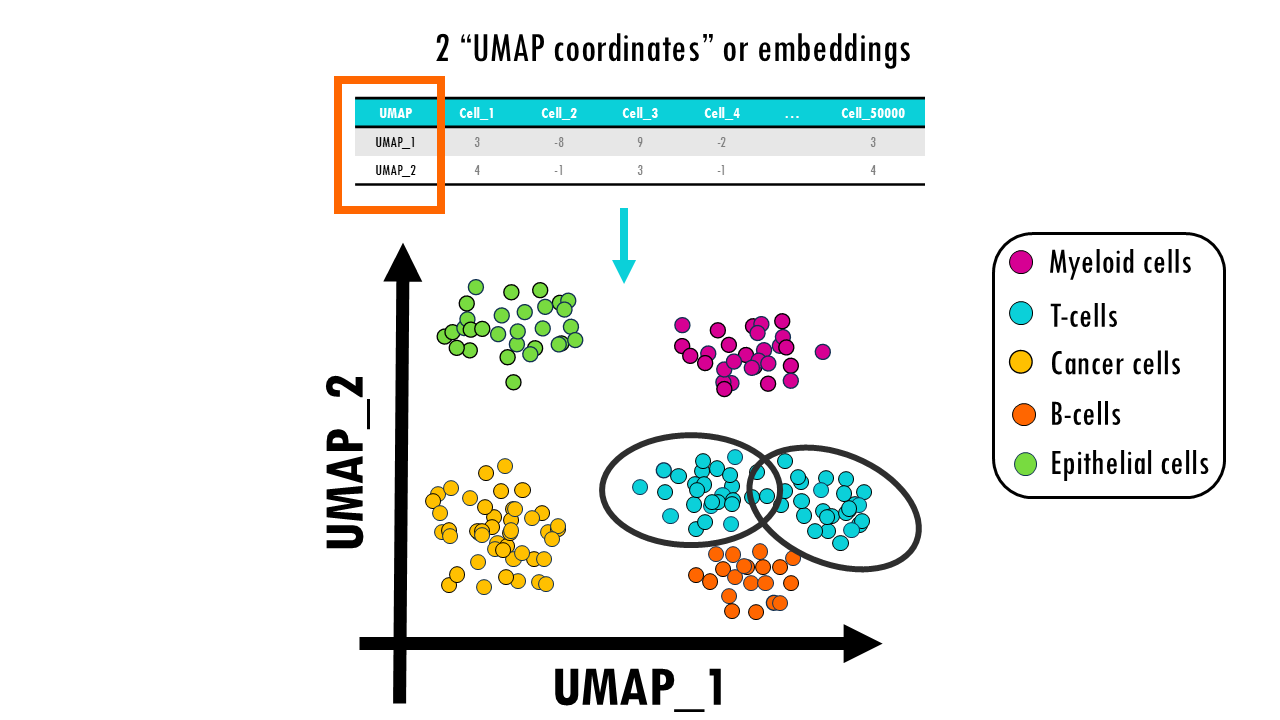
In our snRNAseq dataset we would be able to distinguish 5 main clusters of our main cell types, and you can see how lymphoid cells, B cells and T cells, cluster closer together because they are similar immune cells. We have a nice global organisation of the data, but at the same time, cells with very similar gene expression profiles cluster together.
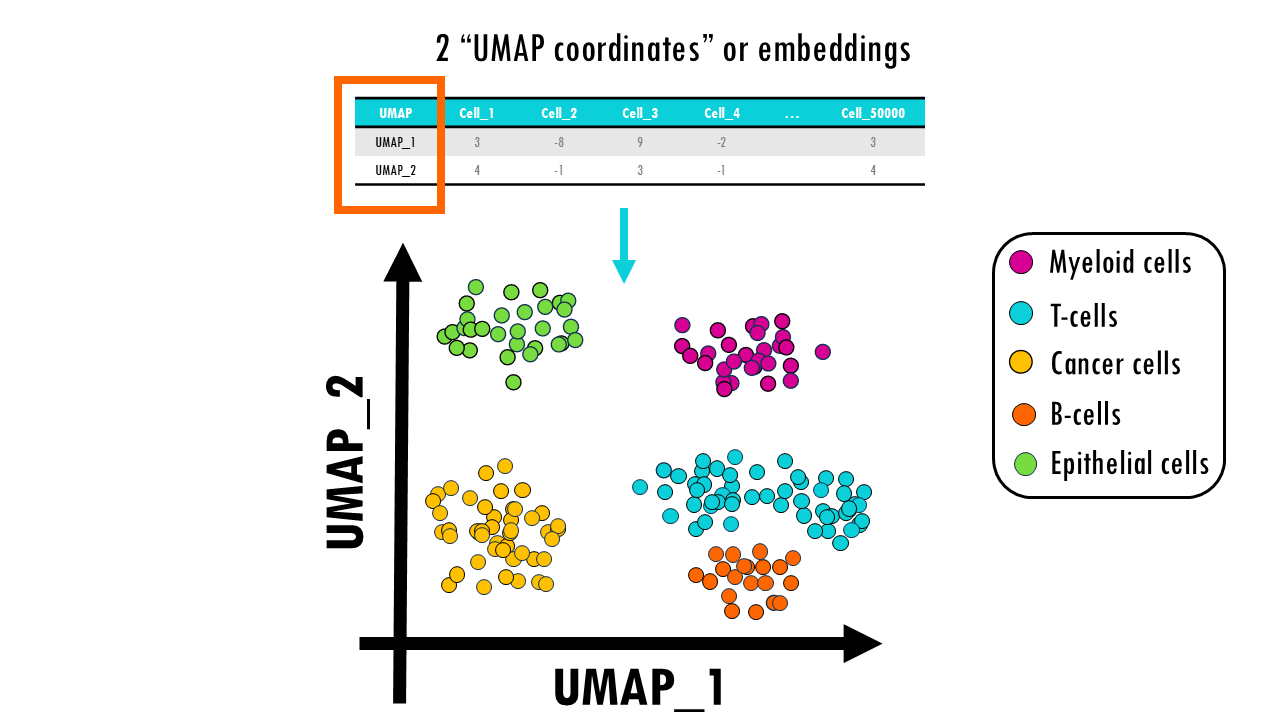
If you look closely enough, you might be able to subcluster the T cell cluster into CD4+ and CD8+ T cells, for example.
How does UMAP work?
So let’s dig in a bit into the maths behind UMAP… without talking about the maths behind UMAP.
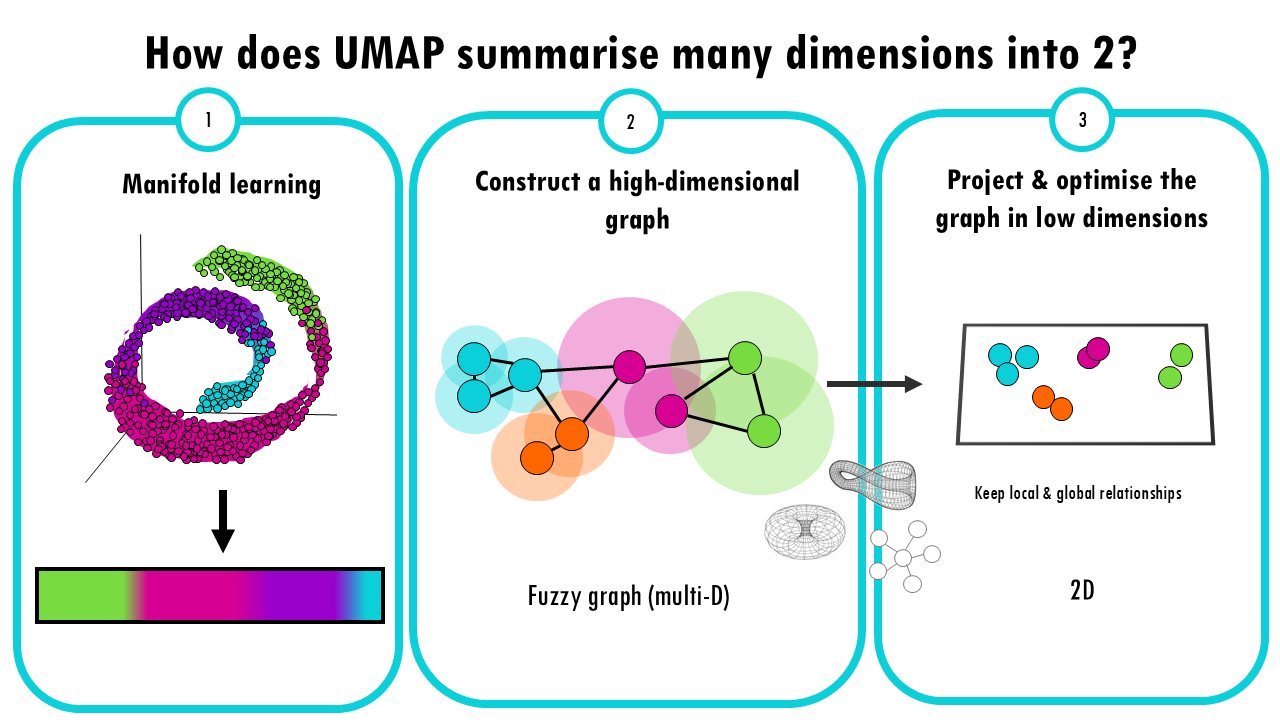
Manifold Learning
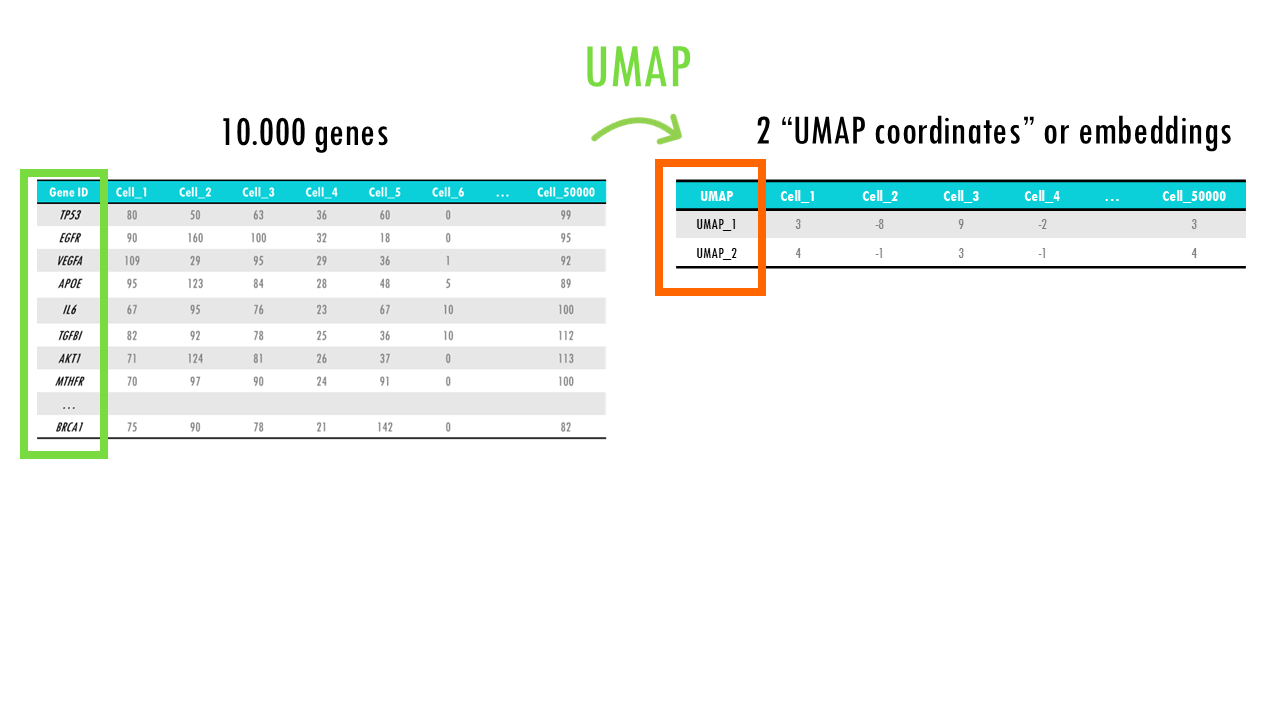
UMAP is based on the idea that high-dimensional data often lies on a lower-dimensional “manifold” (like a curved surface). For example, a 3D object might look like a 2D surface if we zoom in close enough. UMAP tries to learn this manifold structure, but instead of doing it from 3 dimensions to 2, it does it from 10.000 genes to 2 UMAP projections. But the idea is the same.
You can check more about manifold structures here!
Constructing a high-dimensional graph
UMAP starts by creating a “fuzzy graph” which is a high dimensional graph where each data point (in this case, each cell) is connected to its nearest neighbours.
So how does UMAP decide whether two cells are connected? We’ll talk about this high dimensional fuzzy graph in just a bit. But for now, just know that with this multi-dimensional graph, UMAP ensures that local structure is preserved in balance with global structure. This is a key difference and advantage over t-SNE.
Ok, but this fuzzy graph is still highly dimensional, we need to bring it down to 2D.
Optimizing the Graph in Low Dimensions
Again, like t-SNE, there’s an optimization step where UMAP optimizes the layout of a 2D graph trying to keep the connections it computed in the multi-D fuzzy graph. Mathematically it is different to t-SNE: t-SNE uses probability distributions, UMAP uses a technique from topology (the study of shapes) to optimize the graph in a lower-dimensional space.
The idea of this step is to keep the local neighbourhood relationships, the connections between cells, while also trying to preserve the broader, global relationships in the data.
So in essence, UMAP is very similar to t-SNE, they both use algorithms to arrange data in low-dimensional space. This said, UMAP does a better job at finding a balance between emphasizing local versus global structure, and is much faster than t-SNE.
As a result, for each cell in our dataset, we get 2 UMAP embeddings or coordinates.
How does UMAP compute each cell’s nearest neighbours?
Ok, so let’s talk a bit more about that fuzzy graph. How does UMAP construct a high-dimensional fuzzy graph?
First, we position our cells in the multi-dimensional space according to the expression of their 10000 genes. Obviously, we cannot visualise it, but let’s imagine it looks like this:
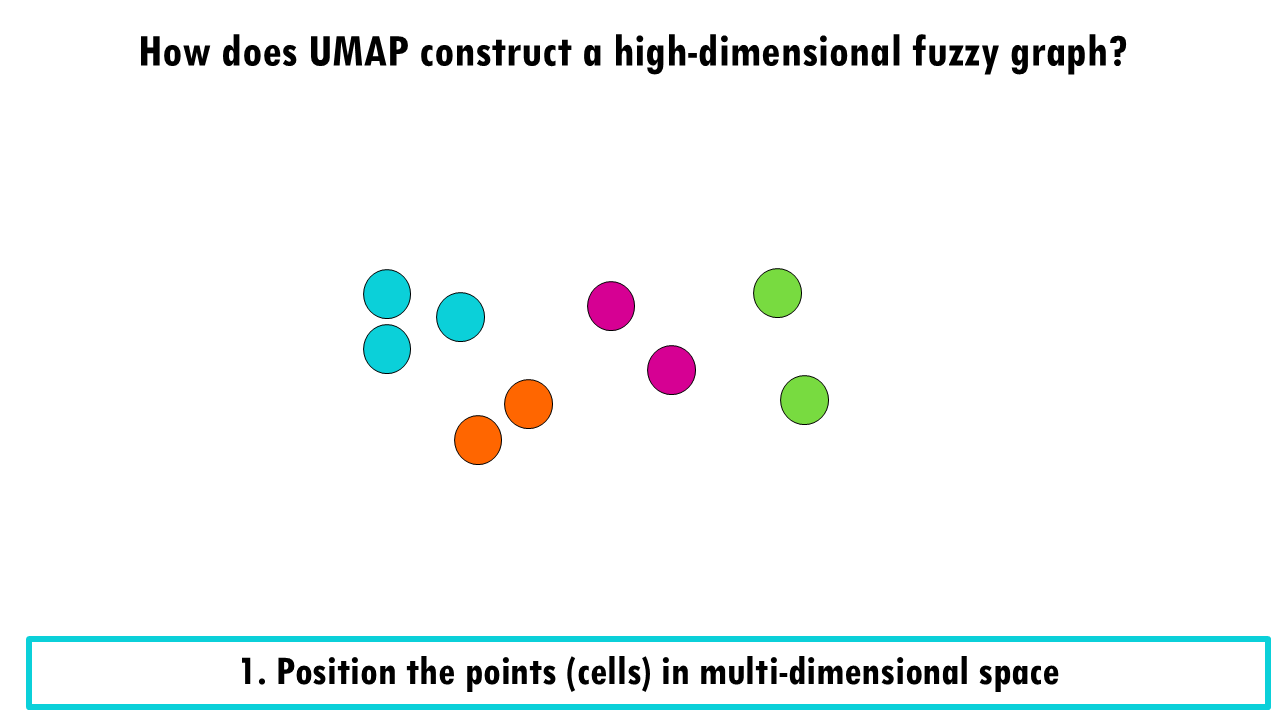
Then, UMAP basically draws a radius from each point or cell and then connects points where there is an overlap.
As you can see, the radius really matters when it comes to deciding whether two cells are connected or not. Notice also that the radius is chosen locally, meaning that each cell has a different radius.
How is the radius chosen? Read more about it below in UMAP’s hyperparameters!
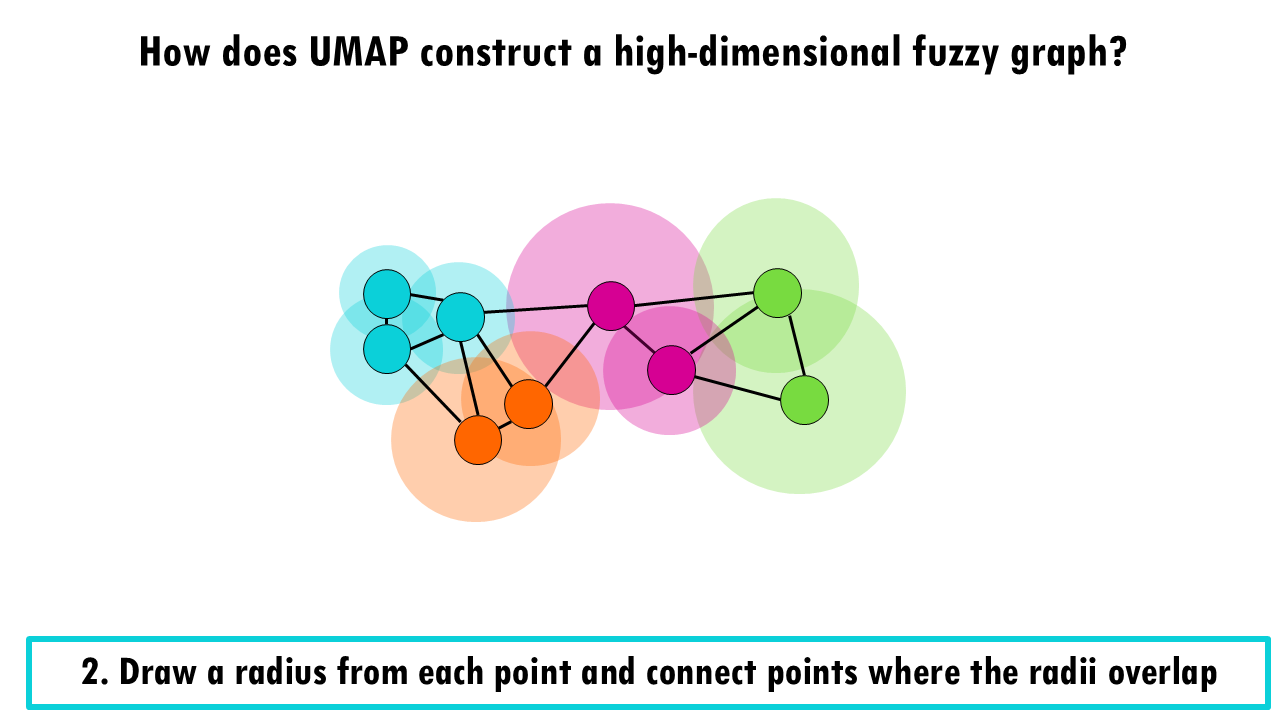
In a third step, UMAP then makes the graph “fuzzy” by decreasing the probability that two cells are connected as the radius grows.
If you take a look at the pink cell in the middle, it’s connecting with high probability with the pink and orange cells which overlap with the smaller radius, and it is connected with lower probability with the blue and green cells which overlap which a much bigger radius.
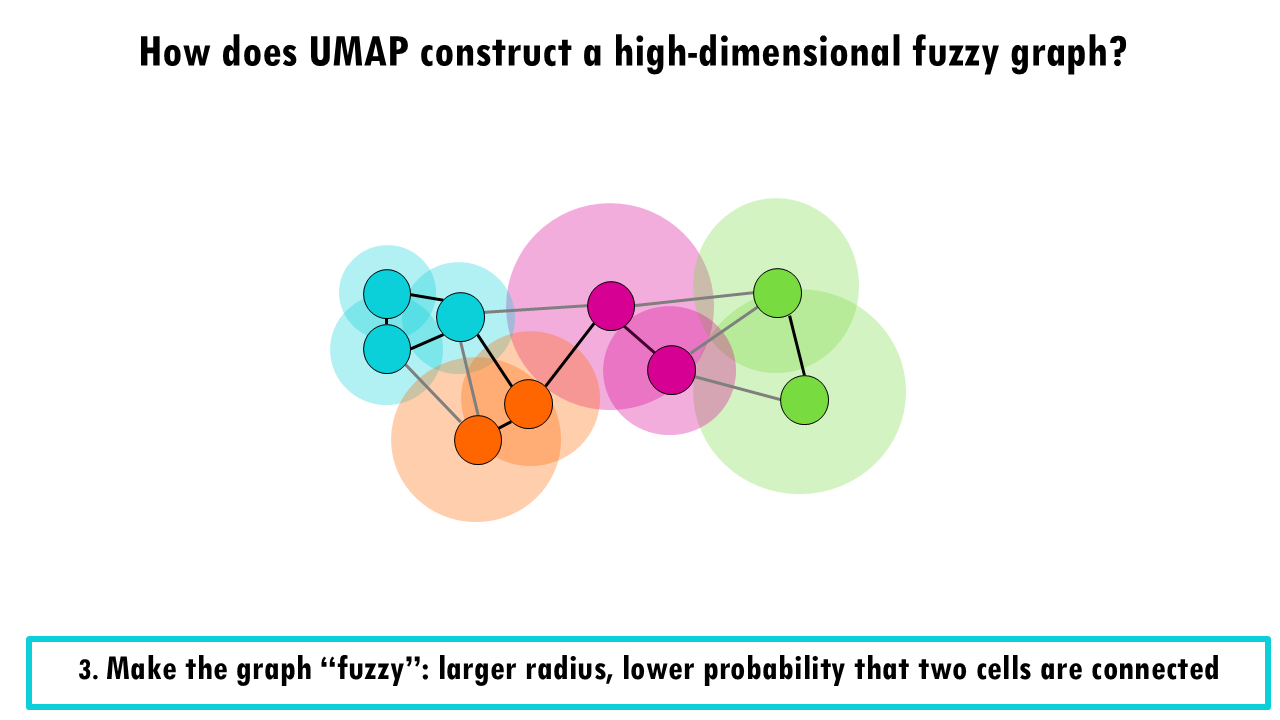
Finally, it makes sure that each point is connected to at least its closest neighbour.
UMAP hyperparameters
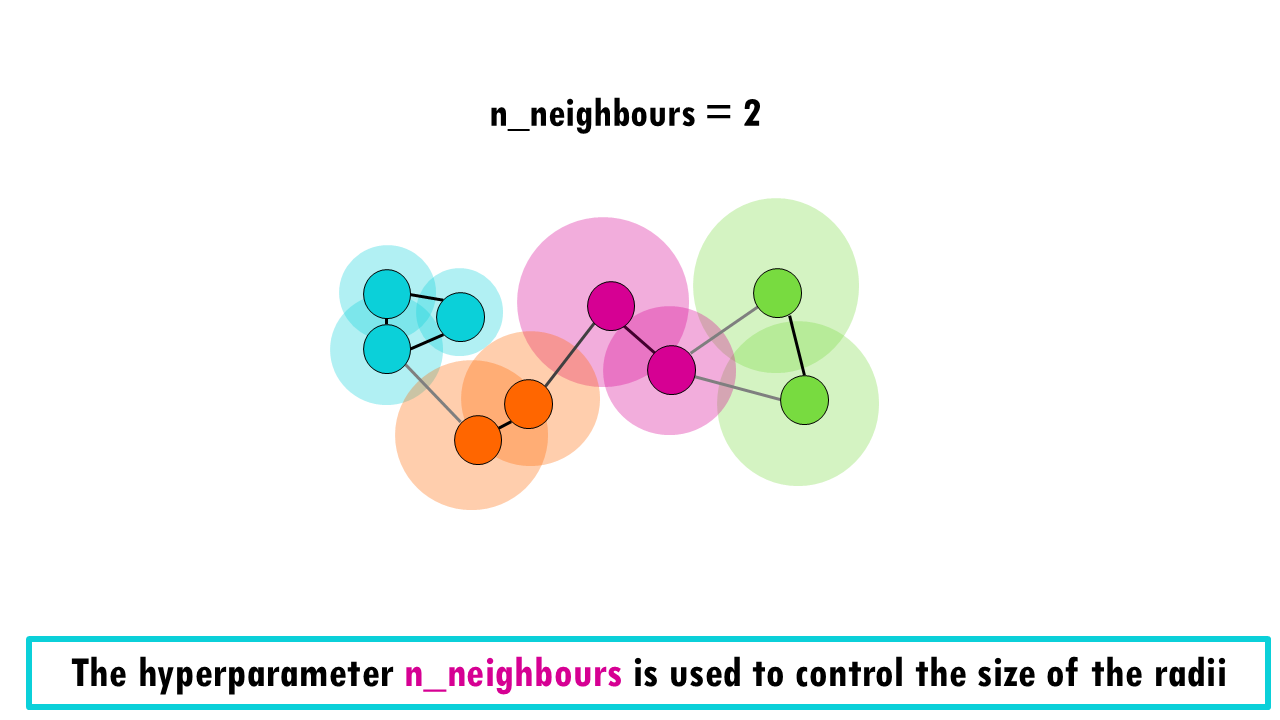
Remember perplexity in t-SNE? Well, UMAP has two hyperparameters that control the results you get. The most important parameter is n_neighbors, which and controls how much of the local versus global structure is preserved.
How does it do that? When building the initial high-dimensional graph, UMAP sets a different radius for each cell to determine which cells are connected vs not. It bases the size of the radii on the distance to each cell’s nearest nth neighbour. This way, all cells are approximately connected to a similar number of neighbouring cells. So for example, if you choose 2 neighbours, the radii would look something like this:
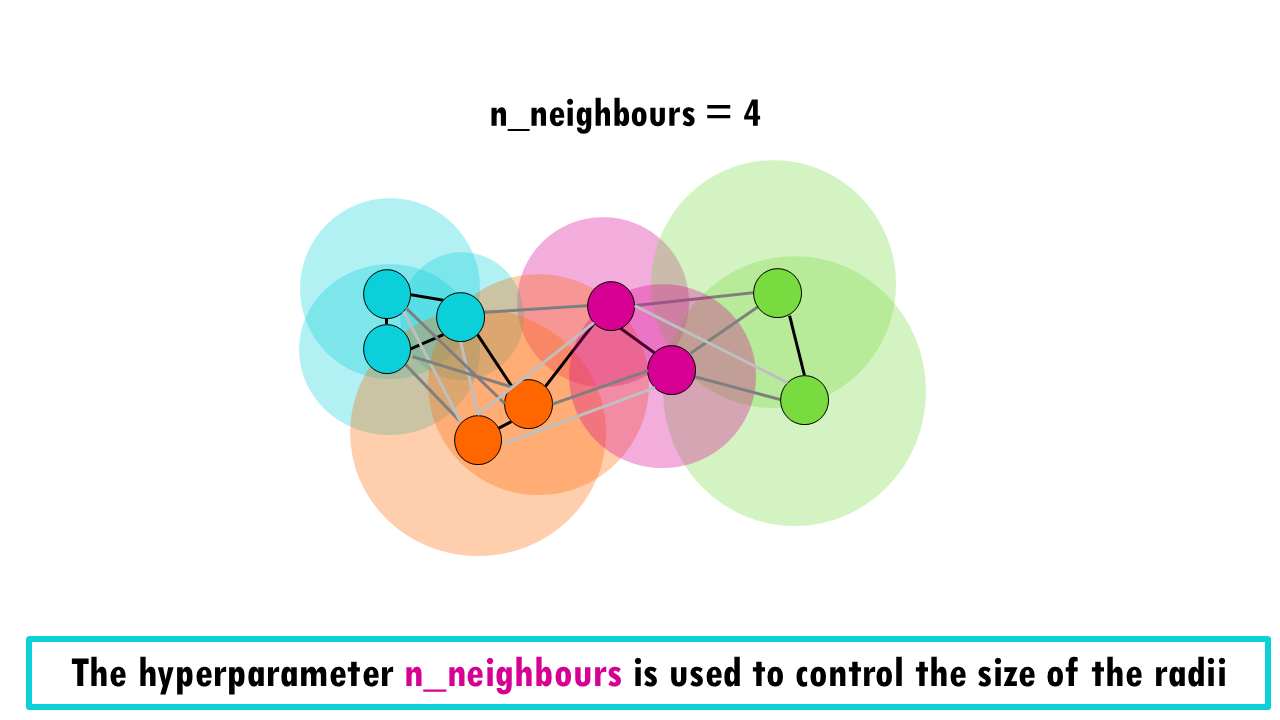
If you set n_neighbours to 4, it might look like this:
In a third step, UMAP then makes the graph “fuzzy” by decreasing the probability that two cells are connected as the radius grows.
If you take a look at the pink cell in the middle, it’s connecting with high probability with the pink and orange cells which overlap with the smaller radius, and it is connected with lower probability with the blue and green cells which overlap which a much bigger radius.
The larger the number of the hyperparameter n_neighbours, the larger radii, so more connections between cells. Notice that the probability of those connections, depicted by the shade of gray, decreases as we connect cells that are further away.
What does this mean in our final plot?
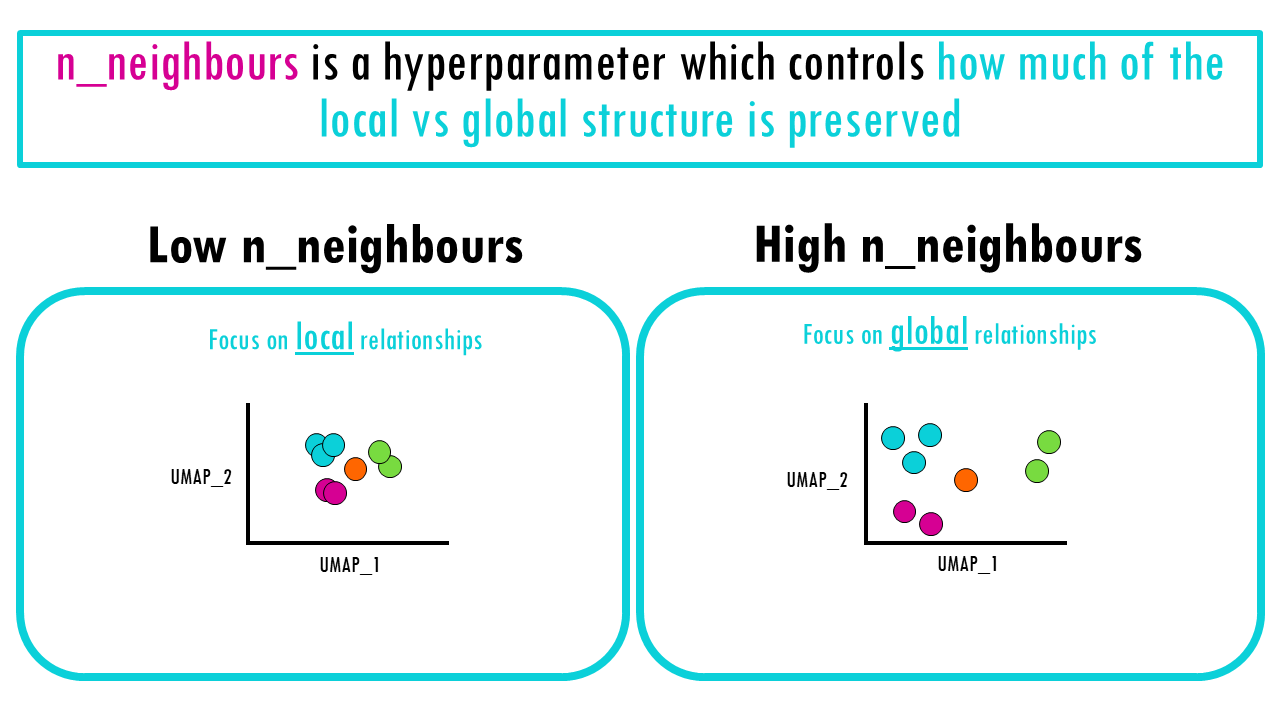
Ultimately, the effect is what we already mention before:
- Low n_neighbours mean local relationships are preserved, meaning cells that are very similar to each other will cluster together.
- High n_neighbours means the clustering might not be as tight, but we focus more on global relationships between cells in our dataset.
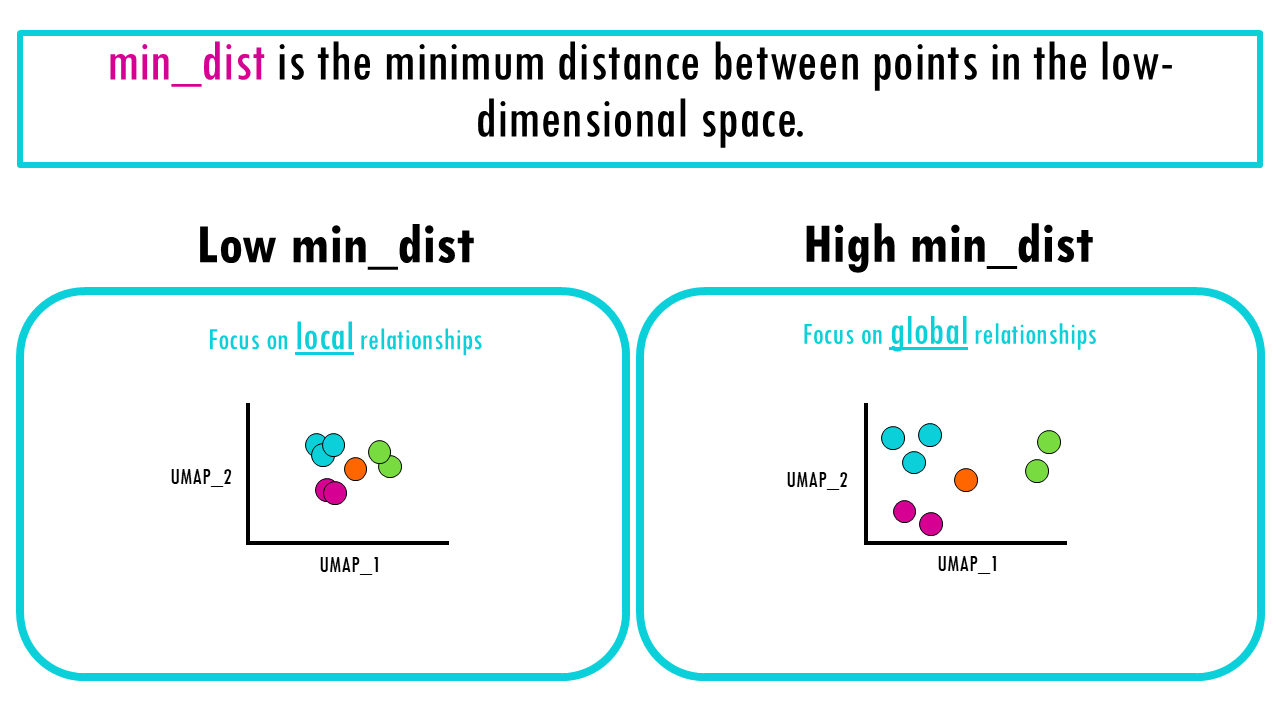
The second parameter you need to be aware of in UMAP is min_dist, which is the minimum distance between points in the low-dimensional space. So we’re not talking about the fuzzy graph anymore, we’re talking about our 2D projection.
The effect has more to do with how you visualise the relationships we already computed. As you can imagine,
- when we reduce the distance between points, UMAP will make cells cluster more tightly together (low min_dist),
- and if you increase min_dist, the cells will space out, so we’re focusing more on the global structure of the dataset.
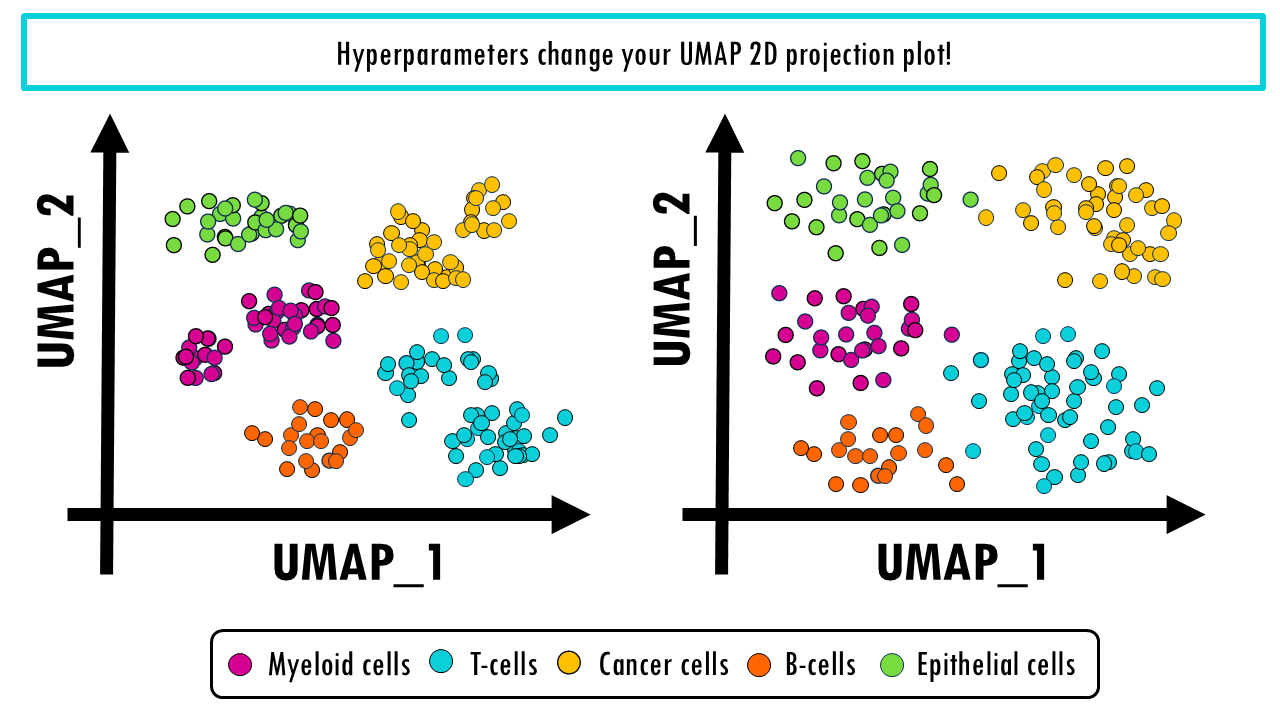
Running UMAP with different parameters can give you very different plots. Which one is right, and which one is wrong? Remember that the purpose of UMAP is to be able to visualise your data, so the number of neighbours allows you to control what is important to you in that visualisation. Do you prefer to be able to distinguish between different T cell subtypes? Or do you prefer a clear general overview of all the cell types in your dataset?
Final notes on UMAP
In summary, UMAP is a dimensionality reduction algorithm which allows us to visualise high-dimensional data in a 2D plot. It does it by modelling each high-dimensional object by a two- or three-dimensional point in such a way that similar objects are modelled by nearby points and dissimilar objects are modelled by distant points. The key maths involves concepts from topology (manifold learning) to build a multi-dimensional fuzzy graph which is then projected to 2D.
However, UMAP also has limitations: the plot you get changes with the values of UMAP’s two main hyperparameters: n_neighbours, and min_dist. So UMAP does require a bit of optimisation to find the right parameters to visualise your dataset.
Nice! So we’ve gone through PCA, t-SNE and UMAP, 3 popular techniques for dimensionality reduction. All 3 methods are great options for our main problem – visualising a large multidimensional dataset with many genes. So, which one to choose? What are the exact differences between them? In part 4 of this series, we will cover differences between PCA, t-SNE and UMAP and when to use each of them.
Want to know more?
Additional resources
If you would like to know more about UMAP, check out:
- UMAP: a really cool webpage which compares t-SNE and UMAP. You can play around with the hyperparameters and how they affect the 2D UMAP projection of a wooly mammoth!
- UMAP: a simple explanation of the key concept in UMAP.
- A deeper dive into the maths behing UMAP.
You might be interested in…
- UMAPs simply explained
- PCA simply explained
- t-SNE simply explained
Ending notes
Wohoo! You made it ’til the end!
In this post, I shared some insights into UMAP and how it works.
Hopefully you found some of my notes and resources useful! Don’t hesitate to leave a comment if there is anything unclear, that you would like explained further, or if you’re looking for more resources on biostatistics! Your feedback is really appreciated and it helps me create more useful content:)
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and… see you in the next one!
Squids don't care much for coffee,
but Laura loves a hot cup in the morning!
If you like my content, you might consider buying me a coffee.
You can also leave a comment or a 'like' in my posts or Youtube channel, knowing that they're helpful really motivates me to keep going:)
Cheers and have a 'squidtastic' day!
And that is the end of this tutorial!
In this post, I explained the differences between log2FC and p-value, and why in differential gene expression analysis we don't always get both high log2FC and low p-value. Hope you found it useful!
Before you go, you might want to check:
Squidtastic!
You made it till the end! Hope you found this post useful.
If you have any questions, or if there are any more topics you would like to see here, leave me a comment down below.
Otherwise, have a very nice day and... see you in the next one!

